You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000151_01000
You are here: Home > Sequence: MGYG000000151_01000
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
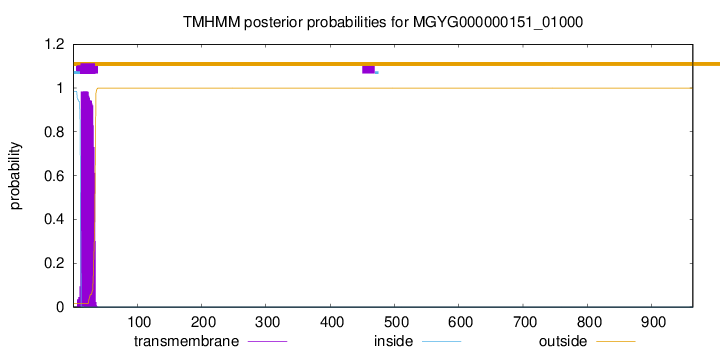
TMHMM annotations
Basic Information help
| Species | Clostridium paraputrificum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Clostridiales; Clostridiaceae; Clostridium; Clostridium paraputrificum | |||||||||||
| CAZyme ID | MGYG000000151_01000 | |||||||||||
| CAZy Family | CBM2 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 386783; End: 389677 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM2 | 871 | 949 | 2.6e-17 | 0.7821782178217822 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00553 | CBM_2 | 1.61e-13 | 871 | 941 | 2 | 74 | Cellulose binding domain. Two tryptophan residues are involved in cellulose binding. Cellulose binding domain found in bacteria. |
| smart00637 | CBD_II | 2.44e-12 | 877 | 941 | 1 | 67 | CBD_II domain. |
| cd02848 | E_set_Chitinase_N | 2.85e-05 | 702 | 757 | 46 | 99 | N-terminal Early set domain associated with the catalytic domain of chitinase. E or "early" set domains are associated with the catalytic domain of chitinase at the N-terminal end. Chitinases hydrolyze the abundant natural biopolymer chitin, producing smaller chito-oligosaccharides. Chitin consists of multiple N-acetyl-D-glucosamine (NAG) residues connected via beta-1,4-glycosidic linkages and is an important structural element of fungal cell wall and arthropod exoskeletons. On the basis of the mode of chitin hydrolysis, chitinases are classified as random, endo-, and exo-chitinases and belong to families 18 and 19 of glycosyl hydrolases based on sequence criteria. The N-terminal domain of chitinase may be related to the immunoglobulin and/or fibronectin type III superfamilies. These domains are associated with different types of catalytic domains at either the N-terminal or C-terminal end and may be involved in homodimeric/tetrameric/dodecameric interactions. Members of this family include members of the alpha amylase family, sialidase, galactose oxidase, cellulase, cellulose, hyaluronate lyase, chitobiase, and chitinase, among others. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QSW17965.1 | 0.0 | 1 | 953 | 1 | 957 |
| QDT41727.1 | 6.17e-198 | 36 | 693 | 160 | 805 |
| QDV49819.1 | 2.58e-197 | 35 | 671 | 166 | 790 |
| QDU09854.1 | 1.46e-192 | 35 | 678 | 158 | 786 |
| QDT98581.1 | 1.47e-191 | 35 | 678 | 158 | 784 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6BT9_A | 4.75e-09 | 770 | 952 | 482 | 660 | ChitinaseChiA74 from Bacillus thuringiensis [Bacillus thuringiensis],6BT9_B Chitinase ChiA74 from Bacillus thuringiensis [Bacillus thuringiensis] |
| 3NDY_E | 8.52e-07 | 868 | 947 | 1 | 82 | Thestructure of the catalytic and carbohydrate binding domain of endoglucanase D from Clostridium cellulovorans [Clostridium cellulovorans],3NDY_F The structure of the catalytic and carbohydrate binding domain of endoglucanase D from Clostridium cellulovorans [Clostridium cellulovorans],3NDY_G The structure of the catalytic and carbohydrate binding domain of endoglucanase D from Clostridium cellulovorans [Clostridium cellulovorans],3NDY_H The structure of the catalytic and carbohydrate binding domain of endoglucanase D from Clostridium cellulovorans [Clostridium cellulovorans],3NDZ_E The structure of the catalytic and carbohydrate binding domain of endoglucanase D from Clostridium cellulovorans bound to cellotriose [Clostridium cellulovorans],3NDZ_F The structure of the catalytic and carbohydrate binding domain of endoglucanase D from Clostridium cellulovorans bound to cellotriose [Clostridium cellulovorans],3NDZ_G The structure of the catalytic and carbohydrate binding domain of endoglucanase D from Clostridium cellulovorans bound to cellotriose [Clostridium cellulovorans],3NDZ_H The structure of the catalytic and carbohydrate binding domain of endoglucanase D from Clostridium cellulovorans bound to cellotriose [Clostridium cellulovorans] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
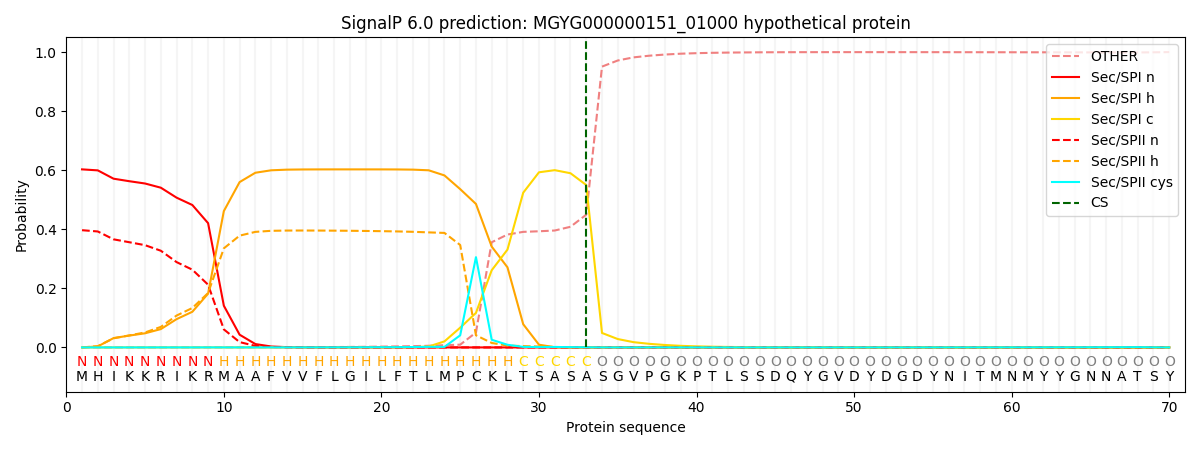
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000633 | 0.596145 | 0.402557 | 0.000262 | 0.000199 | 0.000172 |