You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000151_01522
You are here: Home > Sequence: MGYG000000151_01522
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
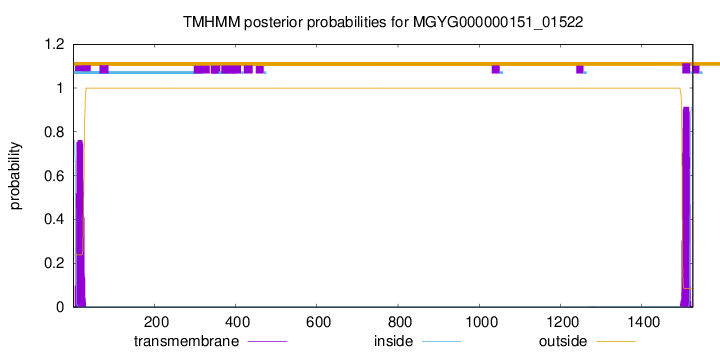
TMHMM annotations
Basic Information help
| Species | Clostridium paraputrificum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Clostridiales; Clostridiaceae; Clostridium; Clostridium paraputrificum | |||||||||||
| CAZyme ID | MGYG000000151_01522 | |||||||||||
| CAZy Family | GH110 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 376141; End: 380721 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH110 | 39 | 604 | 8.1e-145 | 0.9543795620437956 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG1340 | COG1340 | 0.002 | 1338 | 1462 | 59 | 190 | Uncharacterized coiled-coil protein, contains DUF342 domain [Function unknown]. |
| smart01071 | CDC37_N | 0.005 | 1380 | 1472 | 35 | 125 | Cdc37 N terminal kinase binding. Cdc37 is a molecular chaperone required for the activity of numerous eukaryotic protein kinases. This domain corresponds to the N terminal domain which binds predominantly to protein kinases.and is found N terminal to the Hsp (Heat shocked protein) 90-binding domain. Expression of a construct consisting of only the N-terminal domain of Saccharomyces pombe Cdc37 results in cellular viability. This indicates that interactions with the cochaperone Hsp90 may not be essential for Cdc37 function. |
| pfam02368 | Big_2 | 0.009 | 701 | 738 | 24 | 62 | Bacterial Ig-like domain (group 2). This family consists of bacterial domains with an Ig-like fold. Members of this family are found in bacterial and phage surface proteins such as intimins. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AYV35227.1 | 0.0 | 38 | 1469 | 35 | 1446 |
| AGN25168.1 | 0.0 | 38 | 1468 | 35 | 1445 |
| VEH83785.1 | 0.0 | 38 | 1468 | 35 | 1445 |
| QDE01999.1 | 0.0 | 38 | 1468 | 35 | 1445 |
| BAK31224.1 | 0.0 | 38 | 1468 | 35 | 1445 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7JW4_A | 7.15e-42 | 38 | 660 | 27 | 620 | Crystalstructure of PdGH110B in complex with D-galactose [Pseudoalteromonas distincta],7JW4_B Crystal structure of PdGH110B in complex with D-galactose [Pseudoalteromonas distincta] |
| 7JWF_A | 3.08e-41 | 38 | 660 | 27 | 620 | Crystalstructure of PdGH110B D344N in complex with alpha-(1,3)-galactobiose [Pseudoalteromonas distincta],7JWF_B Crystal structure of PdGH110B D344N in complex with alpha-(1,3)-galactobiose [Pseudoalteromonas distincta],7JWF_C Crystal structure of PdGH110B D344N in complex with alpha-(1,3)-galactobiose [Pseudoalteromonas distincta],7JWF_D Crystal structure of PdGH110B D344N in complex with alpha-(1,3)-galactobiose [Pseudoalteromonas distincta] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| B1V8K7 | 8.54e-95 | 38 | 758 | 34 | 723 | Alpha-1,3-galactosidase A OS=Streptacidiphilus griseoplanus OX=66896 GN=glaA PE=1 SV=1 |
| Q826C5 | 2.29e-90 | 37 | 602 | 39 | 587 | Alpha-1,3-galactosidase A OS=Streptomyces avermitilis (strain ATCC 31267 / DSM 46492 / JCM 5070 / NBRC 14893 / NCIMB 12804 / NRRL 8165 / MA-4680) OX=227882 GN=glaA PE=3 SV=1 |
| Q8A2Z5 | 6.54e-76 | 53 | 602 | 23 | 536 | Alpha-1,3-galactosidase A OS=Bacteroides thetaiotaomicron (strain ATCC 29148 / DSM 2079 / JCM 5827 / CCUG 10774 / NCTC 10582 / VPI-5482 / E50) OX=226186 GN=glaA PE=3 SV=1 |
| Q64MU6 | 3.73e-68 | 39 | 602 | 26 | 554 | Alpha-1,3-galactosidase A OS=Bacteroides fragilis (strain YCH46) OX=295405 GN=glaA PE=3 SV=1 |
| Q5L7M8 | 1.69e-67 | 39 | 602 | 26 | 554 | Alpha-1,3-galactosidase A OS=Bacteroides fragilis (strain ATCC 25285 / DSM 2151 / CCUG 4856 / JCM 11019 / NCTC 9343 / Onslow) OX=272559 GN=glaA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
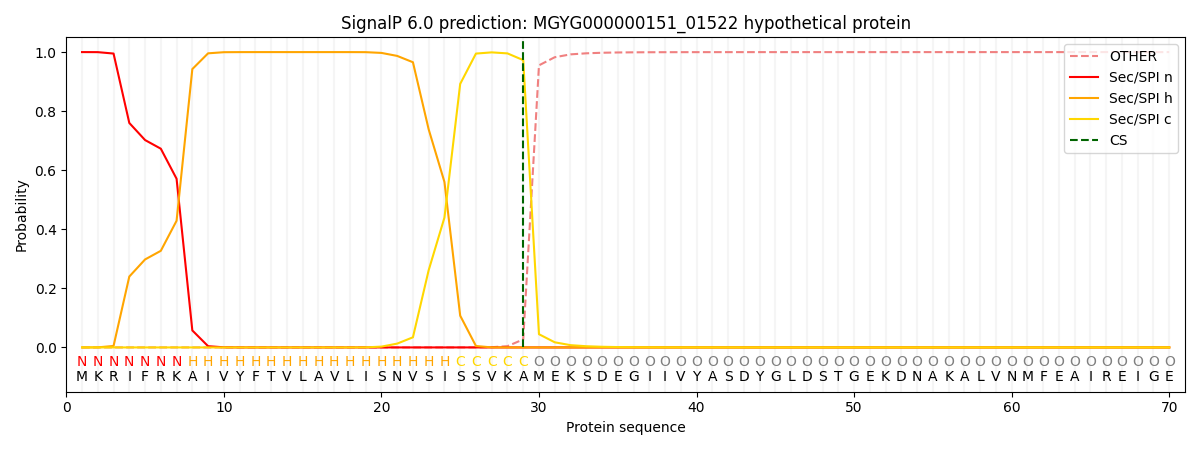
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000275 | 0.999053 | 0.000164 | 0.000178 | 0.000163 | 0.000148 |