You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000151_02705
You are here: Home > Sequence: MGYG000000151_02705
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
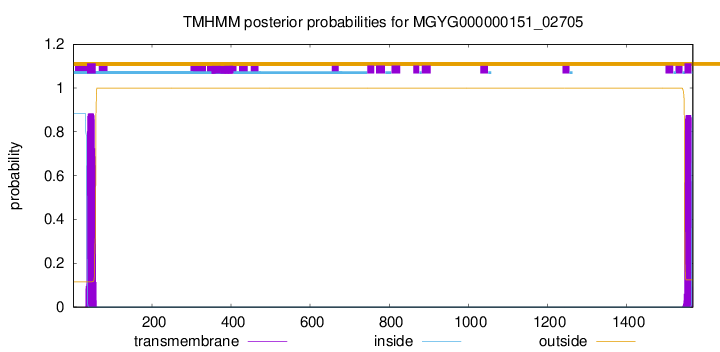
TMHMM annotations
Basic Information help
| Species | Clostridium paraputrificum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Clostridiales; Clostridiaceae; Clostridium; Clostridium paraputrificum | |||||||||||
| CAZyme ID | MGYG000000151_02705 | |||||||||||
| CAZy Family | CBM32 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 136; End: 4845 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM32 | 1257 | 1377 | 9.3e-21 | 0.8870967741935484 |
| CBM32 | 145 | 287 | 1.6e-17 | 0.9193548387096774 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00754 | F5_F8_type_C | 3.61e-17 | 1248 | 1377 | 1 | 127 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| pfam00754 | F5_F8_type_C | 1.91e-08 | 145 | 278 | 3 | 119 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| cd00057 | FA58C | 1.76e-07 | 176 | 274 | 44 | 131 | Substituted updates: Jan 31, 2002 |
| cd00057 | FA58C | 1.07e-05 | 1268 | 1364 | 29 | 130 | Substituted updates: Jan 31, 2002 |
| COG1579 | COG1579 | 0.001 | 1373 | 1505 | 11 | 149 | Predicted nucleic acid-binding protein, contains Zn-ribbon domain [General function prediction only]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AMN35790.1 | 0.0 | 27 | 1513 | 1 | 1493 |
| SQG39183.1 | 0.0 | 27 | 1513 | 1 | 1493 |
| AOY54034.1 | 0.0 | 27 | 1513 | 1 | 1493 |
| BAB80970.1 | 0.0 | 27 | 1513 | 1 | 1493 |
| ABG83191.1 | 0.0 | 27 | 1513 | 1 | 1493 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2V72_A | 5.73e-10 | 1237 | 1379 | 4 | 141 | Thestructure of the family 32 CBM from C. perfringens NanJ in complex with galactose [Clostridium perfringens] |
| 1W8N_A | 5.51e-08 | 1261 | 1377 | 477 | 595 | Contributionof the Active Site Aspartic Acid to Catalysis in the Bacterial Neuraminidase from Micromonospora viridifaciens. [Micromonospora viridifaciens],1W8O_A Contribution of the Active Site Aspartic Acid to Catalysis in the Bacterial Neuraminidase from Micromonospora viridifaciens [Micromonospora viridifaciens] |
| 1WCQ_A | 5.51e-08 | 1261 | 1377 | 477 | 595 | Mutagenesisof the Nucleophilic Tyrosine in a Bacterial Sialidase to Phenylalanine. [Micromonospora viridifaciens],1WCQ_B Mutagenesis of the Nucleophilic Tyrosine in a Bacterial Sialidase to Phenylalanine. [Micromonospora viridifaciens],1WCQ_C Mutagenesis of the Nucleophilic Tyrosine in a Bacterial Sialidase to Phenylalanine. [Micromonospora viridifaciens] |
| 2BER_A | 5.51e-08 | 1261 | 1377 | 477 | 595 | Y370GActive Site Mutant of the Sialidase from Micromonospora viridifaciens in complex with beta-Neu5Ac (sialic acid). [Micromonospora viridifaciens] |
| 2BZD_A | 5.51e-08 | 1261 | 1377 | 477 | 595 | Galactoserecognition by the carbohydrate-binding module of a bacterial sialidase. [Micromonospora viridifaciens],2BZD_B Galactose recognition by the carbohydrate-binding module of a bacterial sialidase. [Micromonospora viridifaciens],2BZD_C Galactose recognition by the carbohydrate-binding module of a bacterial sialidase. [Micromonospora viridifaciens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q02834 | 3.16e-07 | 1261 | 1377 | 523 | 641 | Sialidase OS=Micromonospora viridifaciens OX=1881 GN=nedA PE=1 SV=1 |
| P29767 | 3.82e-07 | 1242 | 1377 | 51 | 181 | Sialidase OS=Clostridium septicum OX=1504 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
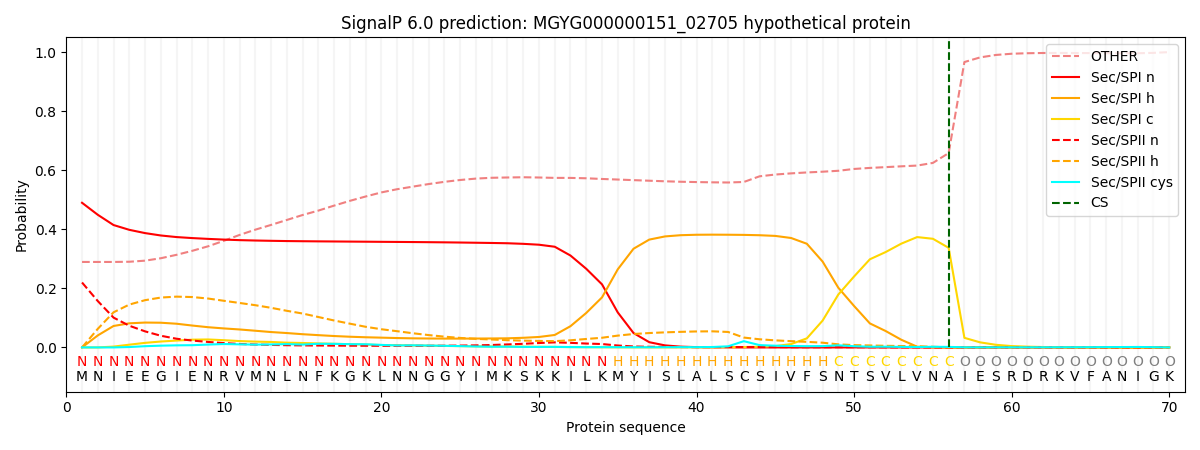
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.296815 | 0.440043 | 0.259620 | 0.001691 | 0.001073 | 0.000754 |