You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000164_01347
You are here: Home > Sequence: MGYG000000164_01347
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Mediterraneibacter_A butyricigenes | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Mediterraneibacter_A; Mediterraneibacter_A butyricigenes | |||||||||||
| CAZyme ID | MGYG000000164_01347 | |||||||||||
| CAZy Family | GH25 | |||||||||||
| CAZyme Description | Cobalamin biosynthesis protein CbiB | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 265594; End: 267369 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH25 | 395 | 578 | 3.1e-46 | 0.9943502824858758 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam03186 | CobD_Cbib | 2.69e-143 | 9 | 300 | 2 | 278 | CobD/Cbib protein. This family includes CobD proteins from a number of bacteria, in Salmonella this protein is called Cbib. Salmonella CobD is a different protein. This protein is involved in cobalamin biosynthesis and is probably an enzyme responsible for the conversion of adenosylcobyric acid to adenosylcobinamide or adenosylcobinamide phosphate. |
| PRK01209 | cobD | 3.31e-135 | 1 | 315 | 1 | 309 | cobalamin biosynthesis protein. |
| COG1270 | CbiB | 3.21e-108 | 1 | 316 | 2 | 314 | Cobalamin biosynthesis protein CobD/CbiB [Coenzyme transport and metabolism]. |
| cd06414 | GH25_LytC-like | 9.25e-87 | 392 | 588 | 1 | 191 | The LytC lysozyme of Streptococcus pneumoniae is a bacterial cell wall hydrolase that cleaves the beta1-4-glycosydic bond located between the N-acetylmuramoyl-N-glucosaminyl residues of the cell wall polysaccharide chains. LytC is composed of a C-terminal glycosyl hydrolase family 25 (GH25) domain and an N-terminal choline-binding module (CBM) consisting of eleven homologous repeats that specifically recognizes the choline residues of pneumococcal lipoteichoic and teichoic acids. This domain arrangement is the reverse of the major pneumococcal autolysin, LytA, and the CPL-1-like lytic enzymes of the pneumococcal bacteriophages, in which the CBM (consisting of six repeats) is at the C-terminus. This model represents the C-terminal catalytic domain of the LytC-like enzymes. |
| TIGR00380 | cobD | 9.81e-67 | 12 | 271 | 11 | 265 | cobalamin biosynthesis protein CobD. This protein is involved in cobalamin (vitamin B12) biosynthesis and porphyrin biosynthesis. It converts cobyric acid to cobinamide by the addition of aminopropanol on the F carboxylic group. It is part of the cob operon. [Biosynthesis of cofactors, prosthetic groups, and carriers, Heme, porphyrin, and cobalamin] |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QRT51228.1 | 5.25e-92 | 345 | 586 | 42 | 282 |
| BBF45127.1 | 2.87e-74 | 333 | 588 | 18 | 277 |
| CBL26148.1 | 9.11e-74 | 348 | 591 | 58 | 305 |
| QRO38911.1 | 4.26e-73 | 307 | 589 | 10 | 286 |
| QBF76167.1 | 4.26e-73 | 307 | 589 | 10 | 286 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4KRU_A | 5.12e-20 | 393 | 590 | 21 | 210 | X-raystructure of catalytic domain of endolysin from clostridium perfringens phage phiSM101 [Clostridium phage phiSM101] |
| 4KRT_A | 3.88e-19 | 393 | 590 | 21 | 210 | X-raystructure of endolysin from clostridium perfringens phage phiSM101 [Clostridium phage phiSM101],4KRT_B X-ray structure of endolysin from clostridium perfringens phage phiSM101 [Clostridium phage phiSM101] |
| 2WAG_A | 4.33e-13 | 393 | 588 | 17 | 207 | TheStructure of a family 25 Glycosyl hydrolase from Bacillus anthracis. [Bacillus anthracis str. Ames] |
| 2WWC_A | 4.81e-12 | 365 | 588 | 238 | 468 | 3D-structureof the modular autolysin LytC from Streptococcus pneumoniae in complex with synthetic peptidoglycan ligand [Streptococcus pneumoniae R6] |
| 1JFX_A | 2.10e-11 | 393 | 583 | 6 | 198 | Crystalstructure of the bacterial lysozyme from Streptomyces coelicolor at 1.65 A resolution [Streptomyces coelicolor] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q8XLK2 | 7.31e-85 | 9 | 314 | 5 | 307 | Cobalamin biosynthesis protein CobD OS=Clostridium perfringens (strain 13 / Type A) OX=195102 GN=cobD PE=3 SV=1 |
| Q7N2S6 | 1.97e-82 | 7 | 317 | 5 | 317 | Cobalamin biosynthesis protein CobD OS=Photorhabdus laumondii subsp. laumondii (strain DSM 15139 / CIP 105565 / TT01) OX=243265 GN=cobD PE=3 SV=1 |
| Q39YE5 | 3.79e-82 | 1 | 314 | 1 | 310 | Cobalamin biosynthesis protein CobD OS=Geobacter metallireducens (strain ATCC 53774 / DSM 7210 / GS-15) OX=269799 GN=cobD PE=3 SV=1 |
| A9MLR0 | 1.98e-78 | 8 | 301 | 7 | 298 | Cobalamin biosynthesis protein CbiB OS=Salmonella arizonae (strain ATCC BAA-731 / CDC346-86 / RSK2980) OX=41514 GN=cbiB PE=3 SV=1 |
| A6TDC6 | 3.91e-78 | 8 | 301 | 7 | 298 | Cobalamin biosynthesis protein CobD OS=Klebsiella pneumoniae subsp. pneumoniae (strain ATCC 700721 / MGH 78578) OX=272620 GN=cobD PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
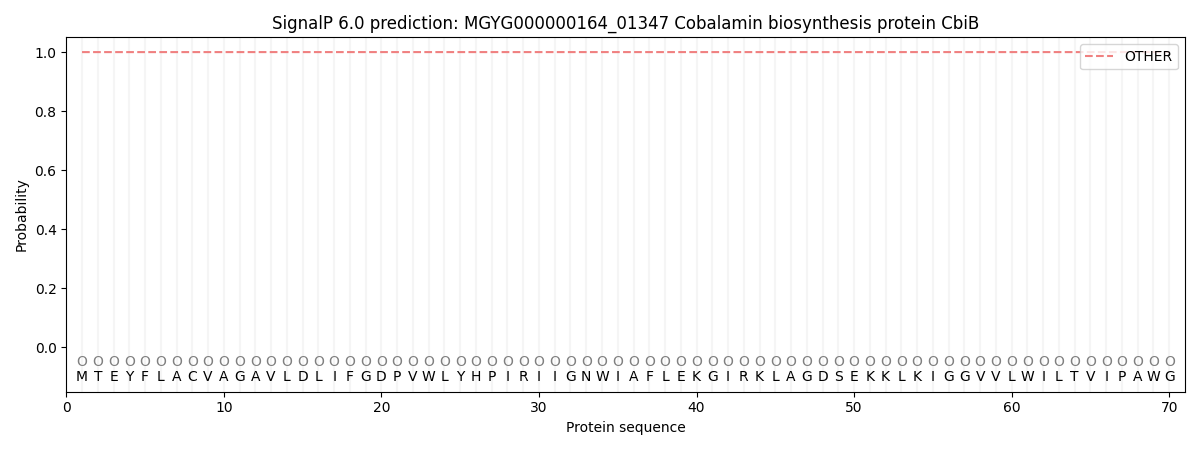
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000029 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |
TMHMM Annotations download full data without filtering help
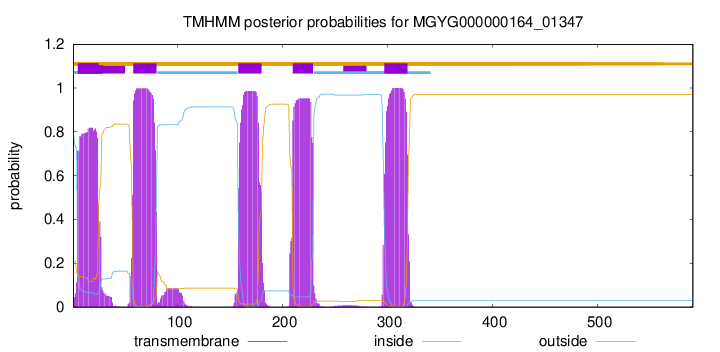
| start | end |
|---|---|
| 5 | 24 |
| 58 | 80 |
| 158 | 180 |
| 210 | 229 |
| 297 | 319 |
