You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000168_00898
You are here: Home > Sequence: MGYG000000168_00898
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
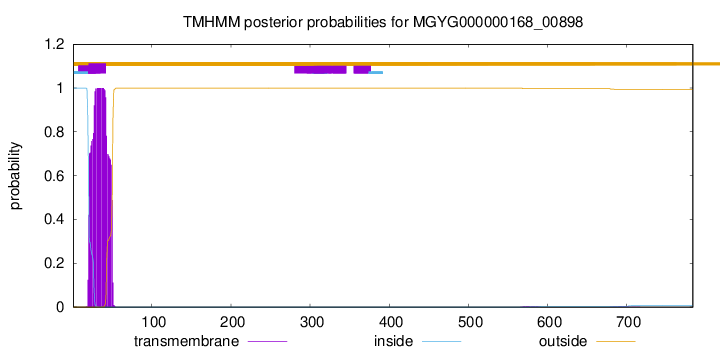
TMHMM annotations
Basic Information help
| Species | Butyricimonas sp003479425 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Marinifilaceae; Butyricimonas; Butyricimonas sp003479425 | |||||||||||
| CAZyme ID | MGYG000000168_00898 | |||||||||||
| CAZy Family | GT51 | |||||||||||
| CAZyme Description | Penicillin-binding protein 1A | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 1094179; End: 1096533 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT51 | 74 | 255 | 8.4e-59 | 0.9943502824858758 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| TIGR02074 | PBP_1a_fam | 1.14e-154 | 85 | 707 | 1 | 530 | penicillin-binding protein, 1A family. Bacterial that synthesize a cell wall of peptidoglycan (murein) generally have several transglycosylases and transpeptidases for the task. This family consists of bifunctional transglycosylase/transpeptidase penicillin-binding proteins (PBP). In the Proteobacteria, this family includes PBP 1A but not the paralogous PBP 1B (TIGR02071). This family also includes related proteins, often designated PBP 1A, from other bacterial lineages. [Cell envelope, Biosynthesis and degradation of murein sacculus and peptidoglycan] |
| COG5009 | MrcA | 2.53e-154 | 22 | 707 | 3 | 720 | Membrane carboxypeptidase/penicillin-binding protein [Cell wall/membrane/envelope biogenesis]. |
| COG0744 | MrcB | 1.68e-141 | 22 | 729 | 15 | 620 | Membrane carboxypeptidase (penicillin-binding protein) [Cell wall/membrane/envelope biogenesis]. |
| PRK11636 | mrcA | 1.01e-92 | 33 | 706 | 3 | 785 | penicillin-binding protein 1a; Provisional |
| TIGR02071 | PBP_1b | 7.32e-86 | 84 | 703 | 145 | 680 | penicillin-binding protein 1B. Bacterial that synthesize a cell wall of peptidoglycan (murein) generally have several transglycosylases and transpeptidases for the task. This family consists of a particular bifunctional transglycosylase/transpeptidase in E. coli and other Proteobacteria, designated penicillin-binding protein 1B. [Cell envelope, Biosynthesis and degradation of murein sacculus and peptidoglycan] |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QRO48595.1 | 0.0 | 1 | 784 | 1 | 784 |
| AZS28569.1 | 0.0 | 17 | 784 | 17 | 784 |
| SNV29696.1 | 0.0 | 29 | 782 | 31 | 780 |
| ADY31956.1 | 0.0 | 29 | 782 | 31 | 780 |
| BAX81636.1 | 5.73e-249 | 36 | 782 | 46 | 797 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4OON_A | 1.79e-80 | 59 | 707 | 12 | 724 | Crystalstructure of PBP1a in complex with compound 17 ((4Z,8S,11E,14S)-5-(2-amino-1,3-thiazol-4-yl)-14-(5,6-dihydroxy-1,3-dioxo-1,3-dihydro-2H-isoindol-2-yl)-8-formyl-2-methyl-6-oxo-3,10-dioxa-4,7,11-triazatetradeca-4,11-diene-2,12,14-tricarboxylic acid) [Pseudomonas aeruginosa PAO1] |
| 3UDF_A | 1.85e-69 | 70 | 717 | 23 | 726 | ChainA, Penicillin-binding protein 1a [Acinetobacter baumannii],3UDF_B Chain B, Penicillin-binding protein 1a [Acinetobacter baumannii],3UDI_A Chain A, Penicillin-binding protein 1a [Acinetobacter baumannii],3UDI_B Chain B, Penicillin-binding protein 1a [Acinetobacter baumannii],3UDX_A Chain A, Penicillin-binding protein 1a [Acinetobacter baumannii],3UDX_B Chain B, Penicillin-binding protein 1a [Acinetobacter baumannii],3UE0_A Chain A, Penicillin-binding protein 1a [Acinetobacter baumannii],3UE0_B Chain B, Penicillin-binding protein 1a [Acinetobacter baumannii],3UE1_A Chain A, Penicillin-binding protein 1a [Acinetobacter baumannii],3UE1_B Chain B, Penicillin-binding protein 1a [Acinetobacter baumannii] |
| 5U2G_A | 1.19e-61 | 52 | 604 | 6 | 568 | 2.6Angstrom Resolution Crystal Structure of Penicillin-Binding Protein 1A from Haemophilus influenzae [Haemophilus influenzae Rd KW20],5U2G_B 2.6 Angstrom Resolution Crystal Structure of Penicillin-Binding Protein 1A from Haemophilus influenzae [Haemophilus influenzae Rd KW20] |
| 3ZG8_B | 1.76e-49 | 175 | 692 | 3 | 450 | CrystalStructure of Penicillin Binding Protein 4 from Listeria monocytogenes in the Ampicillin bound form [Listeria monocytogenes],3ZG9_B Crystal Structure of Penicillin-Binding Protein 4 from Listeria monocytogenes in the Cefuroxime bound form [Listeria monocytogenes],3ZGA_B Crystal Structure of Penicillin-Binding Protein 4 from Listeria monocytogenes in the Carbenicillin bound form [Listeria monocytogenes] |
| 5FGZ_A | 1.10e-46 | 84 | 679 | 154 | 672 | E.coli PBP1b in complex with FPI-1465 [Escherichia coli K-12],5HL9_A E. coli PBP1b in complex with acyl-ampicillin and moenomycin [Escherichia coli K-12],5HLA_A E. coli PBP1b in complex with acyl-cephalexin and moenomycin [Escherichia coli K-12],5HLB_A E. coli PBP1b in complex with acyl-aztreonam and moenomycin [Escherichia coli K-12],5HLD_A E. coli PBP1b in complex with acyl-CENTA and moenomycin [Escherichia coli K-12],6YN0_A Structure of E. coli PBP1b with a FtsN peptide activating transglycosylase activity [Escherichia coli K-12],7LQ6_A Chain A, Penicillin-binding protein 1B [Escherichia coli K-12] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| O66874 | 2.63e-84 | 35 | 723 | 14 | 680 | Penicillin-binding protein 1A OS=Aquifex aeolicus (strain VF5) OX=224324 GN=mrcA PE=1 SV=1 |
| P0A0Z5 | 1.50e-81 | 33 | 707 | 16 | 707 | Penicillin-binding protein 1A OS=Neisseria meningitidis serogroup A / serotype 4A (strain DSM 15465 / Z2491) OX=122587 GN=mrcA PE=3 SV=1 |
| P0A0Z6 | 1.50e-81 | 33 | 707 | 16 | 707 | Penicillin-binding protein 1A OS=Neisseria meningitidis serogroup B (strain MC58) OX=122586 GN=mrcA PE=3 SV=1 |
| O87579 | 1.06e-80 | 34 | 707 | 17 | 707 | Penicillin-binding protein 1A OS=Neisseria lactamica OX=486 GN=mrcA PE=3 SV=1 |
| Q07806 | 1.18e-80 | 52 | 707 | 32 | 751 | Penicillin-binding protein 1A OS=Pseudomonas aeruginosa (strain ATCC 15692 / DSM 22644 / CIP 104116 / JCM 14847 / LMG 12228 / 1C / PRS 101 / PAO1) OX=208964 GN=mrcA PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
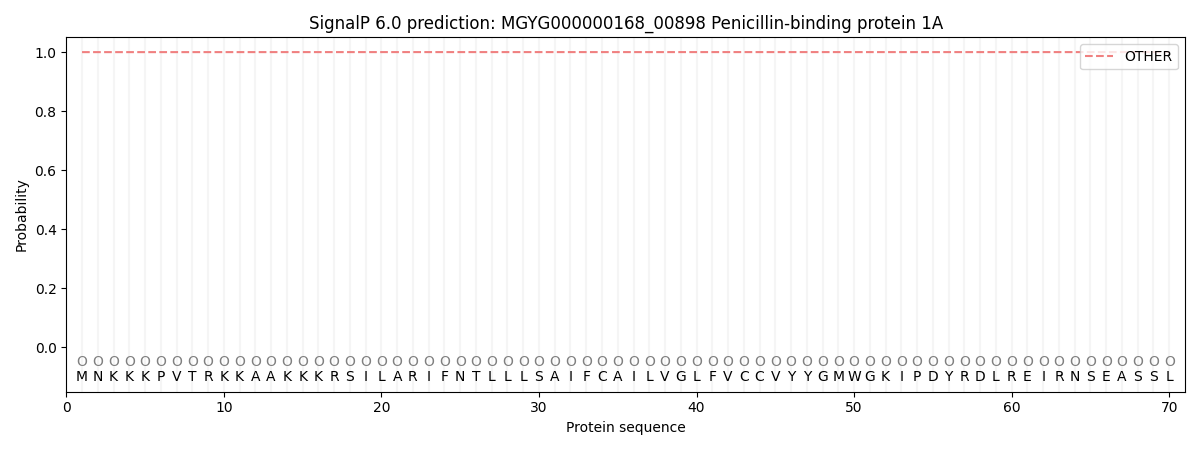
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.999995 | 0.000027 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |