You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000170_00400
You are here: Home > Sequence: MGYG000000170_00400
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Alistipes_A sp900240235 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Rikenellaceae; Alistipes_A; Alistipes_A sp900240235 | |||||||||||
| CAZyme ID | MGYG000000170_00400 | |||||||||||
| CAZy Family | GH2 | |||||||||||
| CAZyme Description | Beta-galactosidase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 461604; End: 464417 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH2 | 20 | 450 | 3.8e-70 | 0.4946808510638298 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3250 | LacZ | 4.51e-21 | 27 | 519 | 12 | 511 | Beta-galactosidase/beta-glucuronidase [Carbohydrate transport and metabolism]. |
| PRK10150 | PRK10150 | 2.17e-19 | 27 | 297 | 12 | 296 | beta-D-glucuronidase; Provisional |
| pfam02837 | Glyco_hydro_2_N | 6.61e-19 | 27 | 158 | 1 | 135 | Glycosyl hydrolases family 2, sugar binding domain. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities and has a jelly-roll fold. The domain binds the sugar moiety during the sugar-hydrolysis reaction. |
| PRK10340 | ebgA | 1.07e-12 | 25 | 432 | 39 | 479 | cryptic beta-D-galactosidase subunit alpha; Reviewed |
| pfam02836 | Glyco_hydro_2_C | 1.67e-04 | 282 | 430 | 4 | 164 | Glycosyl hydrolases family 2, TIM barrel domain. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QGA23712.1 | 0.0 | 1 | 937 | 1 | 937 |
| QOY86859.1 | 6.24e-234 | 58 | 931 | 35 | 864 |
| AQT69276.1 | 2.84e-225 | 2 | 931 | 3 | 918 |
| AQT69150.1 | 3.26e-215 | 22 | 934 | 19 | 942 |
| ASV75422.1 | 7.10e-213 | 23 | 918 | 28 | 941 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6ZJV_A | 1.24e-17 | 29 | 427 | 59 | 467 | Cold-adaptedbeta-D-galactosidase from Arthrobacter sp. 32cB mutant D207A [Arthrobacter sp. 32cB],6ZJW_A Cold-adapted beta-D-galactosidase from Arthrobacter sp. 32cB mutant D207A in complex with galactose [Arthrobacter sp. 32cB],6ZJX_A Cold-adapted beta-D-galactosidase from Arthrobacter sp. 32cB mutant D207A in complex with saccharose [Arthrobacter sp. 32cB] |
| 6LEM_B | 1.28e-17 | 27 | 428 | 10 | 443 | ChainB, Beta-D-glucuronidase [Escherichia coli] |
| 4JHZ_A | 1.29e-17 | 27 | 428 | 14 | 447 | Structureof E. coli beta-Glucuronidase bound with a novel, potent inhibitor 2-[4-(1,3-benzodioxol-5-ylmethyl)piperazin-1-yl]-N-[(1S,2S,5S)-2,5-dimethoxycyclohexyl]acetamide [Escherichia coli K-12],4JHZ_B Structure of E. coli beta-Glucuronidase bound with a novel, potent inhibitor 2-[4-(1,3-benzodioxol-5-ylmethyl)piperazin-1-yl]-N-[(1S,2S,5S)-2,5-dimethoxycyclohexyl]acetamide [Escherichia coli K-12] |
| 6LEJ_B | 1.29e-17 | 27 | 428 | 12 | 445 | ChainB, Beta-D-glucuronidase [Escherichia coli] |
| 6LEG_A | 1.29e-17 | 27 | 428 | 13 | 446 | ChainA, Beta-D-glucuronidase [Escherichia coli],6LEG_B Chain B, Beta-D-glucuronidase [Escherichia coli],6LEG_C Chain C, Beta-D-glucuronidase [Escherichia coli],6LEG_D Chain D, Beta-D-glucuronidase [Escherichia coli],6LEL_A Chain A, Beta-D-glucuronidase [Escherichia coli],6LEL_B Chain B, Beta-D-glucuronidase [Escherichia coli] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P05804 | 7.06e-17 | 27 | 428 | 12 | 445 | Beta-glucuronidase OS=Escherichia coli (strain K12) OX=83333 GN=uidA PE=1 SV=2 |
| Q59140 | 2.17e-12 | 29 | 425 | 31 | 455 | Beta-galactosidase OS=Arthrobacter sp. (strain B7) OX=86041 GN=lacZ PE=1 SV=1 |
| Q56307 | 2.23e-12 | 29 | 404 | 41 | 442 | Beta-galactosidase OS=Thermotoga maritima (strain ATCC 43589 / DSM 3109 / JCM 10099 / NBRC 100826 / MSB8) OX=243274 GN=lacZ PE=1 SV=2 |
| O97524 | 8.52e-12 | 28 | 408 | 39 | 455 | Beta-glucuronidase OS=Felis catus OX=9685 GN=GUSB PE=1 SV=1 |
| Q6LL68 | 3.01e-08 | 28 | 429 | 47 | 484 | Beta-galactosidase OS=Photobacterium profundum (strain SS9) OX=298386 GN=lacZ PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
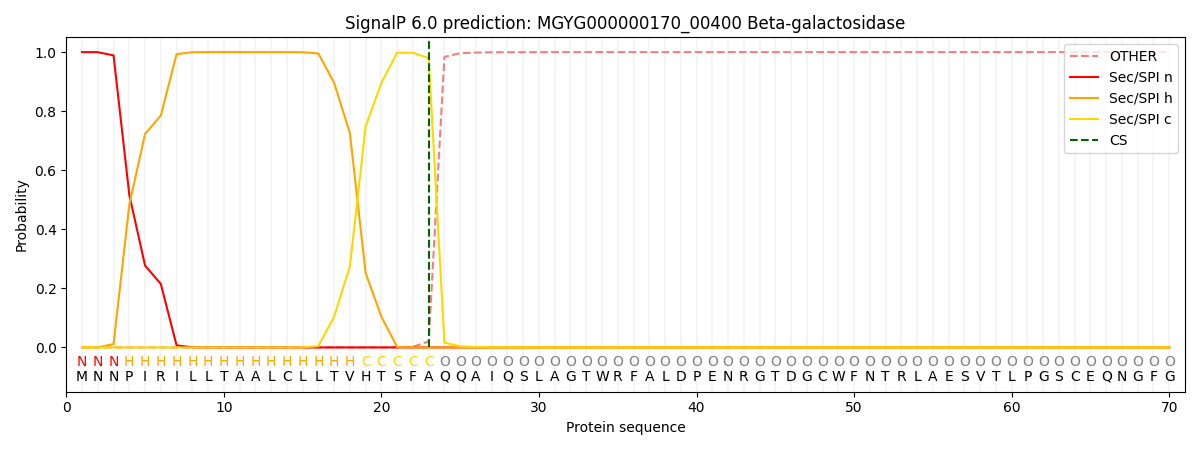
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000231 | 0.999145 | 0.000163 | 0.000157 | 0.000137 | 0.000134 |
