You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000174_01417
You are here: Home > Sequence: MGYG000000174_01417
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Parabacteroides faecis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Tannerellaceae; Parabacteroides; Parabacteroides faecis | |||||||||||
| CAZyme ID | MGYG000000174_01417 | |||||||||||
| CAZy Family | GH2 | |||||||||||
| CAZyme Description | Beta-galactosidase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 28577; End: 31381 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH2 | 18 | 487 | 5.1e-72 | 0.4920212765957447 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3250 | LacZ | 2.27e-27 | 23 | 574 | 12 | 552 | Beta-galactosidase/beta-glucuronidase [Carbohydrate transport and metabolism]. |
| PRK10150 | PRK10150 | 2.67e-22 | 25 | 342 | 14 | 296 | beta-D-glucuronidase; Provisional |
| PRK10340 | ebgA | 5.96e-15 | 9 | 449 | 22 | 455 | cryptic beta-D-galactosidase subunit alpha; Reviewed |
| pfam02837 | Glyco_hydro_2_N | 3.52e-14 | 24 | 216 | 2 | 169 | Glycosyl hydrolases family 2, sugar binding domain. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities and has a jelly-roll fold. The domain binds the sugar moiety during the sugar-hydrolysis reaction. |
| pfam00703 | Glyco_hydro_2 | 2.71e-12 | 219 | 322 | 2 | 106 | Glycosyl hydrolases family 2. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QUT75482.1 | 0.0 | 5 | 934 | 4 | 934 |
| QRO25183.1 | 0.0 | 14 | 931 | 16 | 930 |
| ATP55310.1 | 0.0 | 4 | 932 | 10 | 952 |
| AYB35766.1 | 0.0 | 26 | 933 | 52 | 969 |
| BBE19496.1 | 0.0 | 3 | 931 | 2 | 941 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7SF2_A | 1.83e-15 | 75 | 418 | 46 | 386 | ChainA, Glycosyl hydrolase family 2, sugar binding domain protein [Bacteroides cellulosilyticus DSM 14838],7SF2_B Chain B, Glycosyl hydrolase family 2, sugar binding domain protein [Bacteroides cellulosilyticus DSM 14838],7SF2_C Chain C, Glycosyl hydrolase family 2, sugar binding domain protein [Bacteroides cellulosilyticus DSM 14838],7SF2_D Chain D, Glycosyl hydrolase family 2, sugar binding domain protein [Bacteroides cellulosilyticus DSM 14838],7SF2_E Chain E, Glycosyl hydrolase family 2, sugar binding domain protein [Bacteroides cellulosilyticus DSM 14838],7SF2_F Chain F, Glycosyl hydrolase family 2, sugar binding domain protein [Bacteroides cellulosilyticus DSM 14838] |
| 6D1P_A | 3.34e-15 | 26 | 444 | 17 | 428 | Apostructure of Bacteroides uniformis beta-glucuronidase 3 [Bacteroides uniformis str. 3978 T3 ii],6D1P_B Apo structure of Bacteroides uniformis beta-glucuronidase 3 [Bacteroides uniformis str. 3978 T3 ii] |
| 1BHG_A | 1.77e-14 | 24 | 484 | 19 | 467 | HumanBeta-Glucuronidase At 2.6 A Resolution [Homo sapiens],1BHG_B Human Beta-Glucuronidase At 2.6 A Resolution [Homo sapiens],3HN3_A Human beta-glucuronidase at 1.7 A resolution [Homo sapiens],3HN3_B Human beta-glucuronidase at 1.7 A resolution [Homo sapiens],3HN3_D Human beta-glucuronidase at 1.7 A resolution [Homo sapiens],3HN3_E Human beta-glucuronidase at 1.7 A resolution [Homo sapiens] |
| 3K4A_A | 8.35e-13 | 24 | 342 | 15 | 294 | Crystalstructure of selenomethionine substituted E. coli beta-glucuronidase [Escherichia coli K-12],3K4A_B Crystal structure of selenomethionine substituted E. coli beta-glucuronidase [Escherichia coli K-12] |
| 7KGY_A | 1.11e-12 | 25 | 459 | 19 | 436 | ChainA, Beta-glucuronidase [Faecalibacterium prausnitzii],7KGY_B Chain B, Beta-glucuronidase [Faecalibacterium prausnitzii],7KGY_C Chain C, Beta-glucuronidase [Faecalibacterium prausnitzii],7KGY_D Chain D, Beta-glucuronidase [Faecalibacterium prausnitzii] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q48846 | 3.28e-14 | 9 | 546 | 28 | 560 | Beta-galactosidase large subunit OS=Latilactobacillus sakei OX=1599 GN=lacL PE=3 SV=1 |
| A6TI29 | 3.62e-14 | 18 | 475 | 47 | 490 | Beta-galactosidase 2 OS=Klebsiella pneumoniae subsp. pneumoniae (strain ATCC 700721 / MGH 78578) OX=272620 GN=lacZ2 PE=3 SV=1 |
| A7KGA5 | 4.76e-14 | 18 | 475 | 47 | 490 | Beta-galactosidase OS=Klebsiella pneumoniae OX=573 GN=lacZ PE=3 SV=1 |
| P08236 | 1.03e-13 | 24 | 484 | 39 | 487 | Beta-glucuronidase OS=Homo sapiens OX=9606 GN=GUSB PE=1 SV=2 |
| O77695 | 1.03e-13 | 19 | 480 | 31 | 494 | Beta-glucuronidase (Fragment) OS=Chlorocebus aethiops OX=9534 GN=GUSB PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
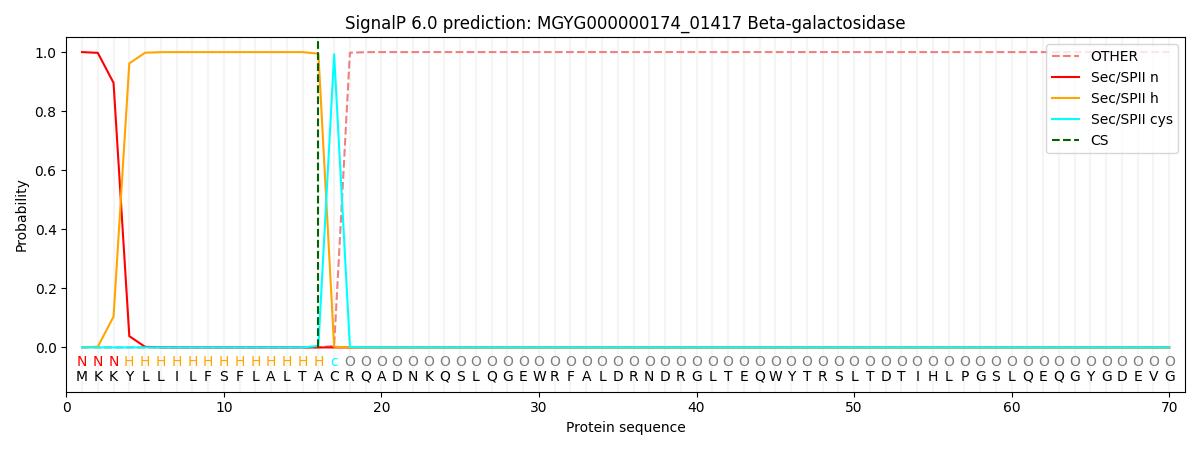
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000032 | 1.000037 | 0.000000 | 0.000000 | 0.000000 |
