You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000174_02516
You are here: Home > Sequence: MGYG000000174_02516
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Parabacteroides faecis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Tannerellaceae; Parabacteroides; Parabacteroides faecis | |||||||||||
| CAZyme ID | MGYG000000174_02516 | |||||||||||
| CAZy Family | GH106 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 187584; End: 190805 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH106 | 344 | 1068 | 8.5e-234 | 0.8859223300970874 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam17132 | Glyco_hydro_106 | 0.0 | 39 | 865 | 1 | 869 | alpha-L-rhamnosidase. |
| cd03143 | A4_beta-galactosidase_middle_domain | 2.40e-04 | 715 | 793 | 53 | 132 | A4 beta-galactosidase middle domain: a type 1 glutamine amidotransferase (GATase1)-like domain. A4 beta-galactosidase middle domain: a type 1 glutamine amidotransferase (GATase1)-like domain. This group includes proteins similar to beta-galactosidase from Thermus thermophilus. Beta-Galactosidase hydrolyzes the beta-1,4-D-galactosidic linkage of lactose, as well as those of related chromogens, o-nitrophenyl-beta-D-galactopyranoside (ONP-Gal) and 5-bromo-4-chloro-3-indolyl-beta-D-galactoside (X-gal). This A4 beta-galactosidase middle domain lacks the catalytic triad of typical GATase1 domains. The reactive Cys residue found in the sharp turn between a beta strand and an alpha helix termed the nucleophile elbow in typical GATase1 domains is not conserved in this group. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QEC53093.1 | 0.0 | 24 | 1067 | 24 | 1074 |
| SCM59077.1 | 0.0 | 33 | 1073 | 34 | 1083 |
| QGA23785.1 | 0.0 | 29 | 1073 | 32 | 1084 |
| ATP59259.1 | 0.0 | 31 | 1067 | 40 | 1085 |
| SCD21026.1 | 0.0 | 34 | 1072 | 33 | 1081 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6Q2F_A | 1.56e-154 | 34 | 1068 | 42 | 1139 | Structureof Rhamnosidase from Novosphingobium sp. PP1Y [Novosphingobium sp. PP1Y] |
| 5MQM_A | 1.89e-118 | 34 | 1066 | 31 | 1097 | Glycosidehydrolase BT_0986 [Bacteroides thetaiotaomicron],5MQN_A Glycoside hydrolase BT_0986 [Bacteroides thetaiotaomicron] |
| 5MWK_A | 5.03e-118 | 34 | 1066 | 31 | 1097 | Glycosidehydrolase BT_0986 [Bacteroides thetaiotaomicron] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| T2KNA8 | 7.31e-147 | 30 | 1067 | 26 | 1155 | Putative beta-glucuronidase OS=Formosa agariphila (strain DSM 15362 / KCTC 12365 / LMG 23005 / KMM 3901 / M-2Alg 35-1) OX=1347342 GN=BN863_22040 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
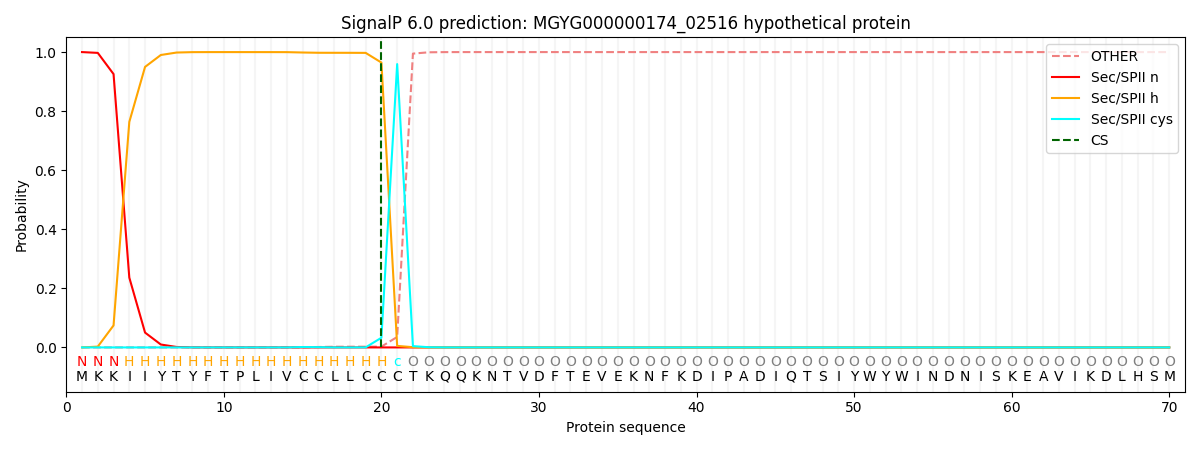
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000016 | 1.000031 | 0.000000 | 0.000000 | 0.000000 |
