You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000174_02574
Basic Information
help
| Species |
Parabacteroides faecis
|
| Lineage |
Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Tannerellaceae; Parabacteroides; Parabacteroides faecis
|
| CAZyme ID |
MGYG000000174_02574
|
| CAZy Family |
GH106 |
| CAZyme Description |
hypothetical protein
|
| CAZyme Property |
|
| Genome Property |
| Genome Assembly ID |
Genome Size |
Genome Type |
Country |
Continent |
| MGYG000000174 |
6515019 |
Isolate |
China |
Asia |
|
| Gene Location |
Start: 288123;
End: 291023
Strand: +
|
No EC number prediction in MGYG000000174_02574.
| Family |
Start |
End |
Evalue |
family coverage |
| GH106 |
186 |
963 |
4.8e-212 |
0.883495145631068 |
| Cdd ID |
Domain |
E-Value |
qStart |
qEnd |
sStart |
sEnd |
Domain Description |
| pfam17132
|
Glyco_hydro_106 |
1.07e-178 |
36 |
753 |
2 |
870 |
alpha-L-rhamnosidase. |
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
Description |
| 6Q2F_A
|
3.58e-110 |
184 |
963 |
407 |
1139 |
Structureof Rhamnosidase from Novosphingobium sp. PP1Y [Novosphingobium sp. PP1Y] |
| 5MQM_A
|
4.13e-100 |
1 |
961 |
1 |
1097 |
Glycosidehydrolase BT_0986 [Bacteroides thetaiotaomicron],5MQN_A Glycoside hydrolase BT_0986 [Bacteroides thetaiotaomicron] |
| 5MWK_A
|
1.08e-99 |
1 |
961 |
1 |
1097 |
Glycosidehydrolase BT_0986 [Bacteroides thetaiotaomicron] |
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
Description |
|
T2KNA8
|
1.11e-125 |
7 |
962 |
3 |
1155 |
Putative beta-glucuronidase OS=Formosa agariphila (strain DSM 15362 / KCTC 12365 / LMG 23005 / KMM 3901 / M-2Alg 35-1) OX=1347342 GN=BN863_22040 PE=2 SV=1 |
This protein is predicted as SP
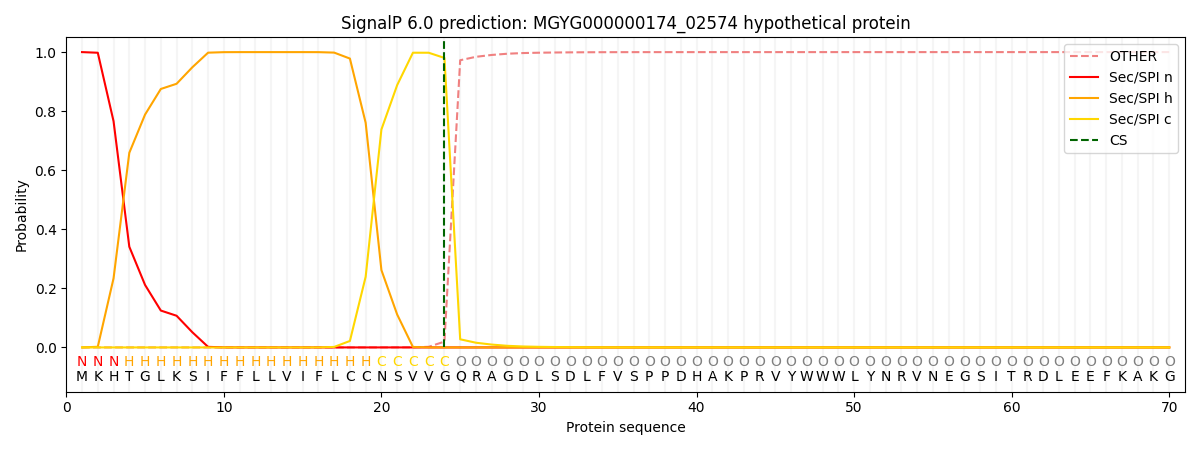
| Other |
SP_Sec_SPI |
LIPO_Sec_SPII |
TAT_Tat_SPI |
TATLIP_Sec_SPII |
PILIN_Sec_SPIII |
|
0.000229
|
0.999211
|
0.000154
|
0.000138
|
0.000120
|
0.000116
|
There is no transmembrane helices in MGYG000000174_02574.

