You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000174_04206
You are here: Home > Sequence: MGYG000000174_04206
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Parabacteroides faecis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Tannerellaceae; Parabacteroides; Parabacteroides faecis | |||||||||||
| CAZyme ID | MGYG000000174_04206 | |||||||||||
| CAZy Family | GH120 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 24374; End: 26278 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH120 | 318 | 409 | 2.1e-36 | 0.989010989010989 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam13229 | Beta_helix | 7.99e-07 | 261 | 409 | 13 | 137 | Right handed beta helix region. This region contains a parallel beta helix region that shares some similarity with Pectate lyases. |
| pfam13229 | Beta_helix | 8.42e-06 | 314 | 448 | 6 | 146 | Right handed beta helix region. This region contains a parallel beta helix region that shares some similarity with Pectate lyases. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ARV16552.1 | 7.58e-223 | 20 | 627 | 19 | 644 |
| AOW11377.1 | 4.80e-217 | 11 | 627 | 3 | 627 |
| SDS19086.1 | 2.40e-211 | 20 | 627 | 24 | 644 |
| QOY88336.1 | 1.48e-209 | 20 | 627 | 45 | 663 |
| AUC24119.1 | 5.94e-197 | 18 | 627 | 1 | 634 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3VST_A | 2.03e-183 | 22 | 627 | 2 | 638 | Thecomplex structure of XylC with Tris [Thermoanaerobacterium saccharolyticum JW/SL-YS485],3VST_B The complex structure of XylC with Tris [Thermoanaerobacterium saccharolyticum JW/SL-YS485],3VST_C The complex structure of XylC with Tris [Thermoanaerobacterium saccharolyticum JW/SL-YS485],3VST_D The complex structure of XylC with Tris [Thermoanaerobacterium saccharolyticum JW/SL-YS485],3VSU_A The complex structure of XylC with xylobiose [Thermoanaerobacterium saccharolyticum JW/SL-YS485],3VSU_B The complex structure of XylC with xylobiose [Thermoanaerobacterium saccharolyticum JW/SL-YS485],3VSU_C The complex structure of XylC with xylobiose [Thermoanaerobacterium saccharolyticum JW/SL-YS485],3VSU_D The complex structure of XylC with xylobiose [Thermoanaerobacterium saccharolyticum JW/SL-YS485],3VSV_A The complex structure of XylC with xylose [Thermoanaerobacterium saccharolyticum JW/SL-YS485],3VSV_B The complex structure of XylC with xylose [Thermoanaerobacterium saccharolyticum JW/SL-YS485],3VSV_C The complex structure of XylC with xylose [Thermoanaerobacterium saccharolyticum JW/SL-YS485],3VSV_D The complex structure of XylC with xylose [Thermoanaerobacterium saccharolyticum JW/SL-YS485] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
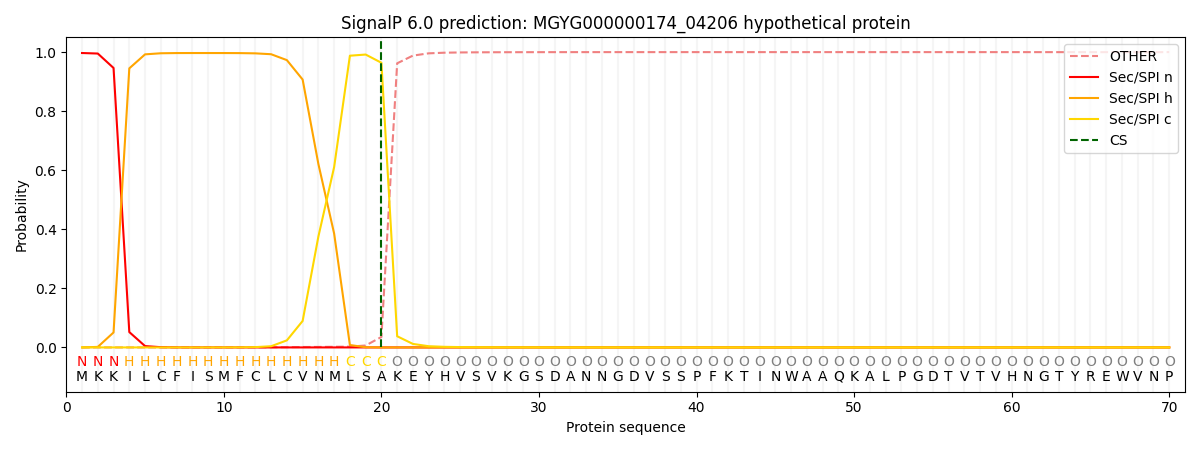
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000463 | 0.994257 | 0.004643 | 0.000225 | 0.000207 | 0.000201 |
