You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000174_04303
You are here: Home > Sequence: MGYG000000174_04303
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
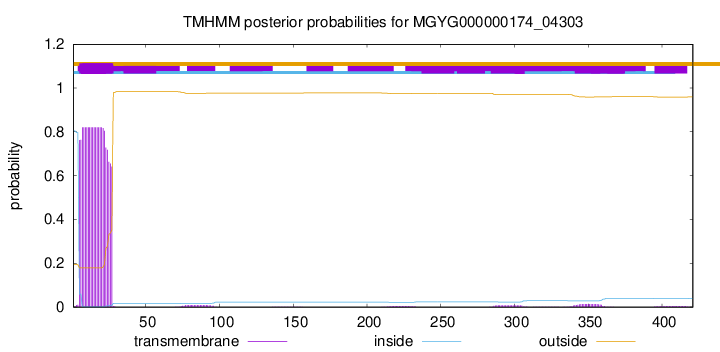
TMHMM annotations
Basic Information help
| Species | Parabacteroides faecis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Tannerellaceae; Parabacteroides; Parabacteroides faecis | |||||||||||
| CAZyme ID | MGYG000000174_04303 | |||||||||||
| CAZy Family | GH76 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 67823; End: 69088 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH76 | 61 | 396 | 5.6e-68 | 0.835195530726257 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam03663 | Glyco_hydro_76 | 1.37e-22 | 73 | 380 | 28 | 308 | Glycosyl hydrolase family 76. Family of alpha-1,6-mannanases. |
| COG4833 | COG4833 | 1.52e-05 | 69 | 358 | 34 | 278 | Predicted alpha-1,6-mannanase, GH76 family [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AAO78630.1 | 5.44e-254 | 1 | 421 | 1 | 420 |
| QQA08345.1 | 5.44e-254 | 1 | 421 | 1 | 420 |
| ALJ44289.1 | 5.44e-254 | 1 | 421 | 1 | 420 |
| QMW85904.1 | 5.44e-254 | 1 | 421 | 1 | 420 |
| QUT69272.1 | 5.44e-254 | 1 | 421 | 1 | 420 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5M77_A | 9.04e-26 | 31 | 420 | 30 | 379 | ChainA, Alpha-1,6-mannanase [Niallia circulans],5M77_B Chain B, Alpha-1,6-mannanase [Niallia circulans] |
| 4D4A_A | 4.17e-25 | 37 | 413 | 23 | 359 | ChainA, Alpha-1,6-mannanase [Niallia circulans],4D4A_B Chain B, Alpha-1,6-mannanase [Niallia circulans],4D4B_A Chain A, Alpha-1,6-mannanase [Niallia circulans],4D4B_B Chain B, Alpha-1,6-mannanase [Niallia circulans],4D4C_A Chain A, Alpha-1,6-mannanase [Niallia circulans],4D4C_B Chain B, Alpha-1,6-mannanase [Niallia circulans],4D4D_A Chain A, Alpha-1,6-mannanase [Niallia circulans],4D4D_B Chain B, Alpha-1,6-mannanase [Niallia circulans],5N0F_A Chain A, Alpha-1,6-mannanase [Niallia circulans],5N0F_B Chain B, Alpha-1,6-mannanase [Niallia circulans],6ZBX_A Chain A, Alpha-1,6-mannanase [Niallia circulans],6ZBX_B Chain B, Alpha-1,6-mannanase [Niallia circulans],7NL5_A Chain A, Alpha-1,6-mannanase [Niallia circulans] |
| 4BOK_A | 4.19e-25 | 37 | 413 | 2 | 338 | ChainA, Alpha-1,6-mannanase [Niallia circulans] |
| 4BOJ_A | 6.00e-25 | 37 | 413 | 5 | 341 | ChainA, Alpha-1,6-mannanase [Niallia circulans],4BOJ_B Chain B, Alpha-1,6-mannanase [Niallia circulans],4BOJ_C Chain C, Alpha-1,6-mannanase [Niallia circulans] |
| 5AGD_A | 1.99e-24 | 37 | 413 | 23 | 359 | ChainA, Alpha-1,6-mannanase [Niallia circulans],5AGD_B Chain B, Alpha-1,6-mannanase [Niallia circulans],6ZBM_A Chain A, Alpha-1,6-mannanase [Niallia circulans],6ZBW_A Chain A, Alpha-1,6-mannanase [Niallia circulans],6ZBW_B Chain B, Alpha-1,6-mannanase [Niallia circulans] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
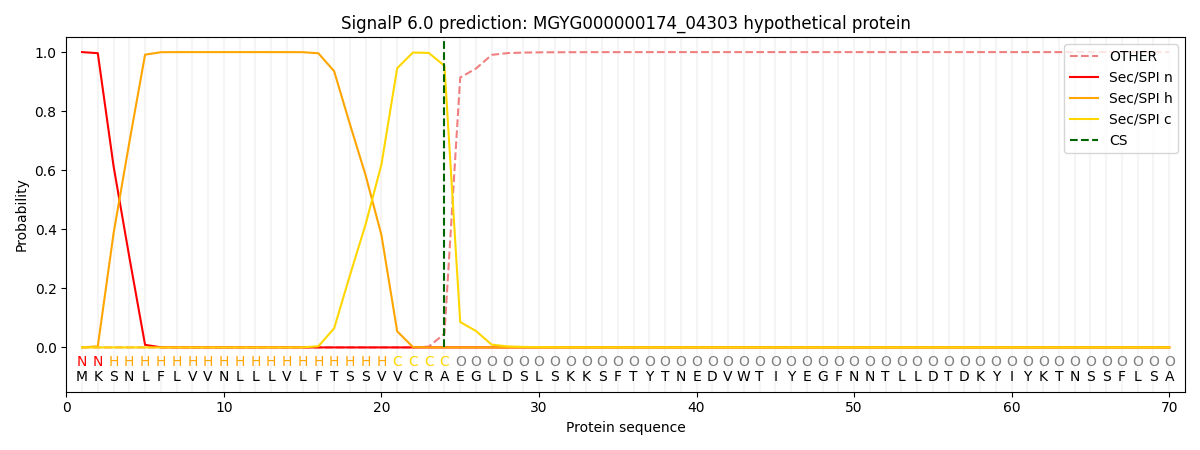
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000181 | 0.999243 | 0.000146 | 0.000156 | 0.000137 | 0.000129 |