You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000174_04708
You are here: Home > Sequence: MGYG000000174_04708
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Parabacteroides faecis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Tannerellaceae; Parabacteroides; Parabacteroides faecis | |||||||||||
| CAZyme ID | MGYG000000174_04708 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 14781; End: 16355 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 40 | 225 | 1.8e-54 | 0.5535714285714286 |
| GH5 | 184 | 421 | 4.8e-18 | 0.5909090909090909 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00150 | Cellulase | 3.67e-22 | 64 | 420 | 23 | 269 | Cellulase (glycosyl hydrolase family 5). |
| COG2730 | BglC | 6.34e-13 | 8 | 420 | 17 | 360 | Aryl-phospho-beta-D-glucosidase BglC, GH1 family [Carbohydrate transport and metabolism]. |
| pfam01301 | Glyco_hydro_35 | 0.003 | 68 | 122 | 27 | 81 | Glycosyl hydrolases family 35. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QRO24956.1 | 9.45e-190 | 11 | 520 | 12 | 521 |
| QNK55040.1 | 2.83e-118 | 31 | 517 | 4 | 491 |
| QJD87094.1 | 1.30e-113 | 31 | 519 | 4 | 494 |
| QTH41332.1 | 1.84e-113 | 31 | 517 | 4 | 492 |
| QDM45840.1 | 5.36e-110 | 31 | 520 | 14 | 503 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2OSW_A | 3.81e-37 | 40 | 482 | 39 | 444 | Endo-glycoceramidaseII from Rhodococcus sp. [Rhodococcus sp.],2OSW_B Endo-glycoceramidase II from Rhodococcus sp. [Rhodococcus sp.],2OYK_A Endo-glycoceramidase II from Rhodococcus sp.: cellobiose-like isofagomine complex [Rhodococcus sp.],2OYK_B Endo-glycoceramidase II from Rhodococcus sp.: cellobiose-like isofagomine complex [Rhodococcus sp.],2OYL_A Endo-glycoceramidase II from Rhodococcus sp.: cellobiose-like imidazole complex [Rhodococcus sp.],2OYL_B Endo-glycoceramidase II from Rhodococcus sp.: cellobiose-like imidazole complex [Rhodococcus sp.],2OYM_A Endo-glycoceramidase II from Rhodococcus sp.: five-membered iminocyclitol complex [Rhodococcus sp.],2OYM_B Endo-glycoceramidase II from Rhodococcus sp.: five-membered iminocyclitol complex [Rhodococcus sp.] |
| 2OSX_A | 1.84e-36 | 40 | 482 | 39 | 444 | ChainA, Endoglycoceramidase II [Rhodococcus sp.] |
| 2OSY_A | 2.52e-36 | 40 | 482 | 39 | 444 | ChainA, Endoglycoceramidase II [Rhodococcus sp.],2OSY_B Chain B, Endoglycoceramidase II [Rhodococcus sp.] |
| 6JYZ_A | 3.82e-24 | 31 | 217 | 25 | 226 | ChainA, Putative secreted endoglycosylceramidase [Rhodococcus hoagii 103S] |
| 5CCU_A | 4.74e-23 | 31 | 477 | 44 | 436 | ChainA, Putative secreted endoglycosylceramidase [Rhodococcus hoagii 103S],5CCU_B Chain B, Putative secreted endoglycosylceramidase [Rhodococcus hoagii 103S] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| A0A3S5YBC7 | 2.47e-22 | 31 | 477 | 36 | 428 | Endoglycoceramidase I OS=Rhodococcus hoagii (strain 103S) OX=685727 GN=REQ_38260 PE=1 SV=1 |
| Q9GV16 | 1.17e-16 | 1 | 217 | 1 | 243 | Endoglycoceramidase OS=Cyanea nozakii OX=135523 PE=1 SV=1 |
| Q6L6S1 | 4.95e-15 | 12 | 218 | 4 | 235 | Endoglycoceramidase OS=Hydra vulgaris OX=6087 PE=1 SV=1 |
| H1AE13 | 1.82e-11 | 31 | 221 | 6 | 255 | Glucosylceramidase OS=Neosartorya fumigata OX=746128 GN=egc1 PE=1 SV=1 |
| I1BTD7 | 1.94e-09 | 35 | 221 | 19 | 262 | Glucosylceramidase OS=Rhizopus delemar (strain RA 99-880 / ATCC MYA-4621 / FGSC 9543 / NRRL 43880) OX=246409 GN=ERC1 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
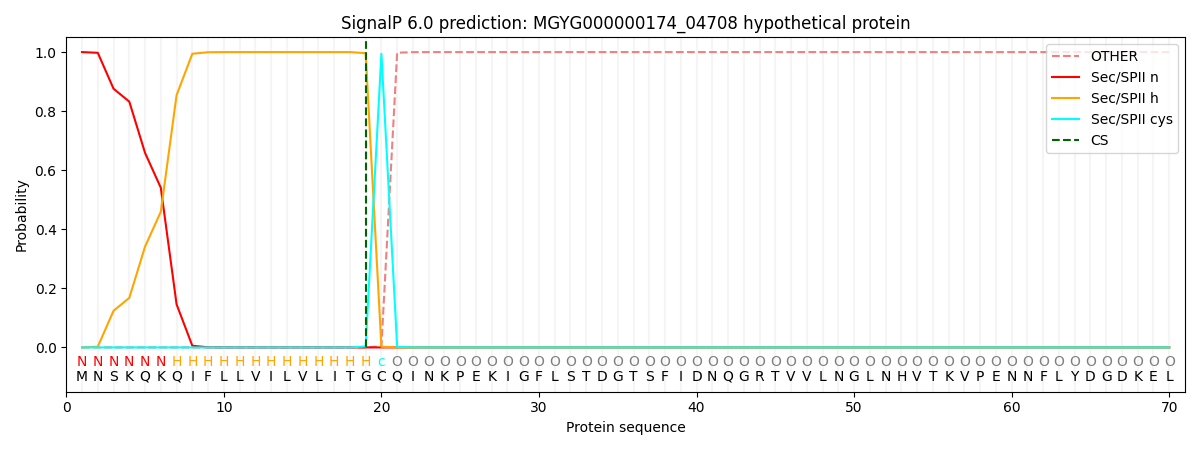
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000000 | 1.000040 | 0.000000 | 0.000000 | 0.000000 |
