You are browsing environment: HUMAN GUT
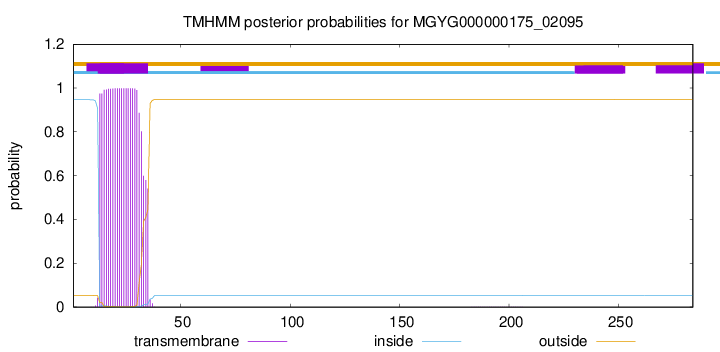
MGYG000000175_02095
Basic Information
help
Species
Muricomes sp000509105
Lineage
Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Muricomes; Muricomes sp000509105
CAZyme ID
MGYG000000175_02095
CAZy Family
GH73
CAZyme Description
hypothetical protein
CAZyme Property
Genome Property
Genome Assembly ID
Genome Size
Genome Type
Country
Continent
MGYG000000175
5055107
Isolate
China
Asia
Gene Location
Start: 57780;
End: 58634
Strand: -
No EC number prediction in MGYG000000175_02095.
Family
Start
End
Evalue
family coverage
GH73
149
279
9.9e-19
0.96875
Cdd ID
Domain
E-Value
qStart
qEnd
sStart
sEnd
Domain Description
NF000535
MSCRAMM_SdrC
3.15e-05
6
120
18
125
MSCRAMM family adhesin SdrC. Features of this protein family include a YSIRK-type signal peptide at the N-terminus and a variable-length C-terminal region of Ser-Asp (SD) repeats followed by an LPXTG motif for surface immobilization by sortase.
more
PTZ00121
PTZ00121
0.008
38
136
1642
1741
MAEBL; Provisional
more
Hit ID
E-Value
Query Start
Query End
Hit Start
Hit End
Description
O51481
8.89e-19
145
248
68
171
Uncharacterized protein BB_0531 OS=Borreliella burgdorferi (strain ATCC 35210 / DSM 4680 / CIP 102532 / B31) OX=224326 GN=BB_0531 PE=3 SV=1
more
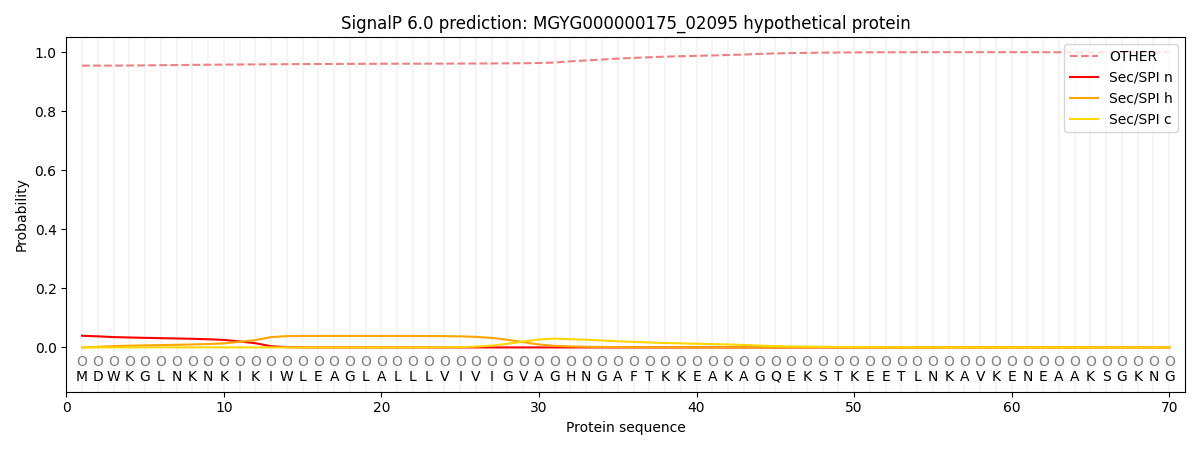
This protein is predicted as OTHER
Other
SP_Sec_SPI
LIPO_Sec_SPII
TAT_Tat_SPI
TATLIP_Sec_SPII
PILIN_Sec_SPIII
0.956163
0.036768
0.000649
0.000159
0.000123
0.006166