You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000183_01081
You are here: Home > Sequence: MGYG000000183_01081
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Butyricimonas paravirosa | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Marinifilaceae; Butyricimonas; Butyricimonas paravirosa | |||||||||||
| CAZyme ID | MGYG000000183_01081 | |||||||||||
| CAZy Family | GH84 | |||||||||||
| CAZyme Description | O-GlcNAcase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 118110; End: 120323 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH84 | 149 | 441 | 1.3e-110 | 0.9932203389830508 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam07555 | NAGidase | 9.73e-165 | 149 | 440 | 1 | 293 | beta-N-acetylglucosaminidase. This family has previously been described as a hyaluronidase. However, more recently it has been shown that this family has beta-N-acetylglucosaminidase activity. |
| pfam18344 | CBM32 | 8.07e-18 | 605 | 670 | 1 | 64 | Carbohydrate binding module family 32. This domain is found in GH84C present in Clostridium perfringens. GH84C is a beta-N-acetylglucosaminidase. This domain is a family 32 carbohydrate binding module (CBM) which preferentially recognizes the non-reducing terminus of N-acetyllactosamine. |
| pfam02838 | Glyco_hydro_20b | 2.46e-14 | 22 | 142 | 1 | 123 | Glycosyl hydrolase family 20, domain 2. This domain has a zincin-like fold. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QRO48327.1 | 0.0 | 1 | 737 | 1 | 737 |
| AZS29316.1 | 0.0 | 1 | 736 | 1 | 736 |
| QQT77482.1 | 0.0 | 20 | 731 | 21 | 730 |
| ASM65960.1 | 0.0 | 20 | 731 | 21 | 730 |
| QRP57274.1 | 0.0 | 20 | 731 | 21 | 730 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2VVN_A | 1.15e-315 | 2 | 731 | 4 | 730 | BtGH84in complex with NH-Butylthiazoline [Bacteroides thetaiotaomicron VPI-5482],2VVN_B BtGH84 in complex with NH-Butylthiazoline [Bacteroides thetaiotaomicron VPI-5482],2VVS_A BtGH84 structure in complex with PUGNAc [Bacteroides thetaiotaomicron VPI-5482],2X0H_A BtGH84 Michaelis complex [Bacteroides thetaiotaomicron VPI-5482],2X0H_B BtGH84 Michaelis complex [Bacteroides thetaiotaomicron VPI-5482],4AIS_A A complex structure of BtGH84 [Bacteroides thetaiotaomicron VPI-5482],4AIS_B A complex structure of BtGH84 [Bacteroides thetaiotaomicron VPI-5482],7OU8_AAA Chain AAA, O-GlcNAcase BT_4395 [Bacteroides thetaiotaomicron VPI-5482],7OU8_BBB Chain BBB, O-GlcNAcase BT_4395 [Bacteroides thetaiotaomicron VPI-5482] |
| 7K41_A | 4.88e-315 | 20 | 731 | 3 | 712 | ChainA, O-GlcNAcase BT_4395 [Escherichia coli K-12] |
| 2J47_A | 6.21e-315 | 21 | 731 | 1 | 709 | Bacteroidesthetaiotaomicron GH84 O-GlcNAcase in complex with a imidazole-pugnac hybrid inhibitor [Bacteroides thetaiotaomicron VPI-5482],2W4X_A BtGH84 in complex with STZ [Bacteroides thetaiotaomicron VPI-5482],2W66_A BtGH84 in complex with HQ602 [Bacteroides thetaiotaomicron VPI-5482],2W66_B BtGH84 in complex with HQ602 [Bacteroides thetaiotaomicron VPI-5482],2W67_A BtGH84 in complex with FMA34 [Bacteroides thetaiotaomicron VPI-5482],2W67_B BtGH84 in complex with FMA34 [Bacteroides thetaiotaomicron VPI-5482],2WCA_A BtGH84 in complex with n-butyl pugnac [Bacteroides thetaiotaomicron VPI-5482],2XJ7_A BtGH84 in complex with 6-acetamido-6-deoxy-castanospermine [Bacteroides thetaiotaomicron VPI-5482],2XJ7_B BtGH84 in complex with 6-acetamido-6-deoxy-castanospermine [Bacteroides thetaiotaomicron VPI-5482],2XM1_A BtGH84 in complex with N-acetyl gluconolactam [Bacteroides thetaiotaomicron VPI-5482],2XM1_B BtGH84 in complex with N-acetyl gluconolactam [Bacteroides thetaiotaomicron VPI-5482],2XM2_A BtGH84 in complex with LOGNAc [Bacteroides thetaiotaomicron VPI-5482],2XM2_B BtGH84 in complex with LOGNAc [Bacteroides thetaiotaomicron VPI-5482],4UR9_A Structure of ligand bound glycosylhydrolase [Bacteroides thetaiotaomicron],4UR9_B Structure of ligand bound glycosylhydrolase [Bacteroides thetaiotaomicron],5FKY_A Structure of a hydrolase bound with an inhibitor [Bacteroides thetaiotaomicron],5FKY_B Structure of a hydrolase bound with an inhibitor [Bacteroides thetaiotaomicron],5FL0_A Structure of a hydrolase with an inhibitor [Bacteroides thetaiotaomicron],5FL0_B Structure of a hydrolase with an inhibitor [Bacteroides thetaiotaomicron],5FL1_A Structure of a hydrolase with an inhibitor [Bacteroides thetaiotaomicron],5FL1_B Structure of a hydrolase with an inhibitor [Bacteroides thetaiotaomicron] |
| 2WZH_A | 6.58e-315 | 2 | 731 | 4 | 730 | BtGH84D242N in complex with MeUMB-derived oxazoline [Bacteroides thetaiotaomicron VPI-5482],4AIU_A A complex structure of BtGH84 [Bacteroides thetaiotaomicron VPI-5482] |
| 2WZI_A | 6.58e-315 | 2 | 731 | 4 | 730 | BtGH84D243N in complex with 5F-oxazoline [Bacteroides thetaiotaomicron VPI-5482],2WZI_B BtGH84 D243N in complex with 5F-oxazoline [Bacteroides thetaiotaomicron VPI-5482] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q89ZI2 | 6.27e-315 | 2 | 731 | 4 | 730 | O-GlcNAcase BT_4395 OS=Bacteroides thetaiotaomicron (strain ATCC 29148 / DSM 2079 / JCM 5827 / CCUG 10774 / NCTC 10582 / VPI-5482 / E50) OX=226186 GN=BT_4395 PE=1 SV=1 |
| Q8XL08 | 2.08e-108 | 27 | 545 | 48 | 583 | O-GlcNAcase NagJ OS=Clostridium perfringens (strain 13 / Type A) OX=195102 GN=nagJ PE=1 SV=1 |
| Q0TR53 | 4.03e-108 | 27 | 545 | 48 | 583 | O-GlcNAcase NagJ OS=Clostridium perfringens (strain ATCC 13124 / DSM 756 / JCM 1290 / NCIMB 6125 / NCTC 8237 / Type A) OX=195103 GN=nagJ PE=1 SV=1 |
| P26831 | 4.27e-54 | 70 | 559 | 97 | 619 | Hyaluronoglucosaminidase OS=Clostridium perfringens (strain 13 / Type A) OX=195102 GN=nagH PE=1 SV=2 |
| O60502 | 3.54e-44 | 150 | 420 | 63 | 340 | Protein O-GlcNAcase OS=Homo sapiens OX=9606 GN=OGA PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
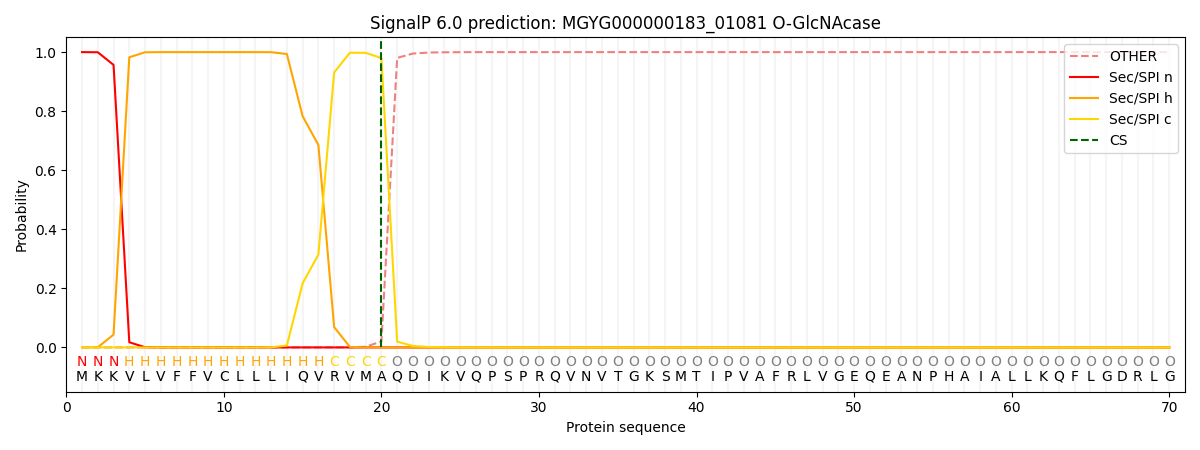
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000189 | 0.999185 | 0.000150 | 0.000159 | 0.000145 | 0.000140 |
