You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000187_00475
You are here: Home > Sequence: MGYG000000187_00475
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
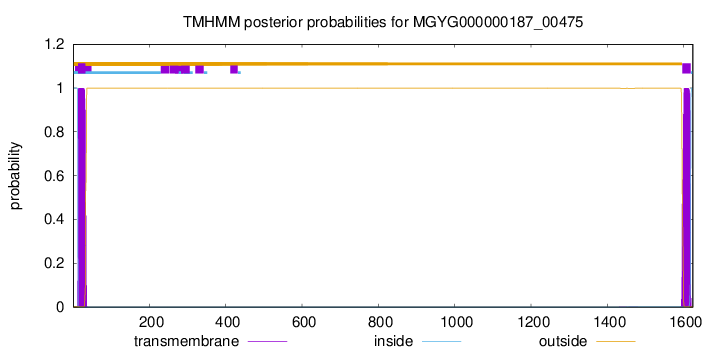
TMHMM annotations
Basic Information help
| Species | Lachnospira sp003537285 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Lachnospira; Lachnospira sp003537285 | |||||||||||
| CAZyme ID | MGYG000000187_00475 | |||||||||||
| CAZy Family | PL1 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 14946; End: 19820 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| PL9 | 825 | 1245 | 4.4e-111 | 0.976 |
| PL1 | 238 | 410 | 2.5e-45 | 0.8267326732673267 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3866 | PelB | 6.63e-47 | 226 | 414 | 82 | 279 | Pectate lyase [Carbohydrate transport and metabolism]. |
| smart00656 | Amb_all | 6.72e-32 | 229 | 412 | 3 | 190 | Amb_all domain. |
| pfam00544 | Pec_lyase_C | 7.19e-21 | 204 | 408 | 1 | 211 | Pectate lyase. This enzyme forms a right handed beta helix structure. Pectate lyase is an enzyme involved in the maceration and soft rotting of plant tissue. |
| pfam04886 | PT | 1.29e-08 | 1481 | 1515 | 2 | 36 | PT repeat. This short repeat is composed on the tetrapeptide XPTX. This repeat is found in a variety of proteins, however it is not clear if these repeats are homologous to each other. The alignment represents nine copies of this repeat. |
| pfam04886 | PT | 1.18e-06 | 1485 | 1519 | 2 | 36 | PT repeat. This short repeat is composed on the tetrapeptide XPTX. This repeat is found in a variety of proteins, however it is not clear if these repeats are homologous to each other. The alignment represents nine copies of this repeat. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ACR72246.1 | 0.0 | 1 | 1624 | 1 | 1623 |
| ACR72247.1 | 0.0 | 1 | 1477 | 1 | 1285 |
| QQY81127.1 | 1.61e-237 | 44 | 1166 | 24 | 1179 |
| AUS06374.1 | 3.54e-235 | 44 | 1133 | 24 | 1151 |
| ADL51369.1 | 6.84e-229 | 42 | 1235 | 57 | 1227 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1RU4_A | 2.31e-34 | 849 | 1131 | 40 | 293 | ChainA, Pectate lyase [Dickeya chrysanthemi] |
| 3VMV_A | 2.24e-20 | 204 | 410 | 47 | 248 | Crystalstructure of pectate lyase Bsp165PelA from Bacillus sp. N165 [Bacillus sp. N16-5],3VMW_A Crystal structure of pectate lyase Bsp165PelA from Bacillus sp. N165 in complex with trigalacturonate [Bacillus sp. N16-5] |
| 5OLQ_A | 7.45e-20 | 856 | 1138 | 35 | 327 | Rhamnogalacturonanlyase [Bacteroides thetaiotaomicron],5OLQ_B Rhamnogalacturonan lyase [Bacteroides thetaiotaomicron],5OLQ_C Rhamnogalacturonan lyase [Bacteroides thetaiotaomicron],5OLR_A Rhamnogalacturonan lyase [Bacteroides thetaiotaomicron],5OLR_B Rhamnogalacturonan lyase [Bacteroides thetaiotaomicron],5OLR_C Rhamnogalacturonan lyase [Bacteroides thetaiotaomicron],5OLS_A Rhamnogalacturonan lyase [Bacteroides thetaiotaomicron] |
| 1VBL_A | 7.66e-18 | 196 | 408 | 93 | 330 | Structureof the thermostable pectate lyase PL 47 [Bacillus sp. TS-47] |
| 3ZSC_A | 3.56e-16 | 227 | 397 | 54 | 225 | Catalyticfunction and substrate recognition of the pectate lyase from Thermotoga maritima [Thermotoga maritima] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P0C1A6 | 5.58e-35 | 837 | 1131 | 53 | 318 | Pectate lyase L OS=Dickeya chrysanthemi OX=556 GN=pelL PE=3 SV=1 |
| P0C1A7 | 2.02e-33 | 849 | 1131 | 65 | 318 | Pectate lyase L OS=Dickeya dadantii (strain 3937) OX=198628 GN=pelL PE=1 SV=1 |
| P22751 | 6.86e-25 | 829 | 1141 | 353 | 646 | Pectate disaccharide-lyase OS=Dickeya chrysanthemi OX=556 GN=pelX PE=1 SV=1 |
| Q8GCB2 | 4.02e-22 | 184 | 417 | 64 | 281 | Pectate trisaccharide-lyase OS=Bacillus licheniformis OX=1402 GN=pelA PE=1 SV=1 |
| Q65DC2 | 4.02e-22 | 184 | 417 | 64 | 281 | Pectate trisaccharide-lyase OS=Bacillus licheniformis (strain ATCC 14580 / DSM 13 / JCM 2505 / CCUG 7422 / NBRC 12200 / NCIMB 9375 / NCTC 10341 / NRRL NRS-1264 / Gibson 46) OX=279010 GN=BLi04129 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
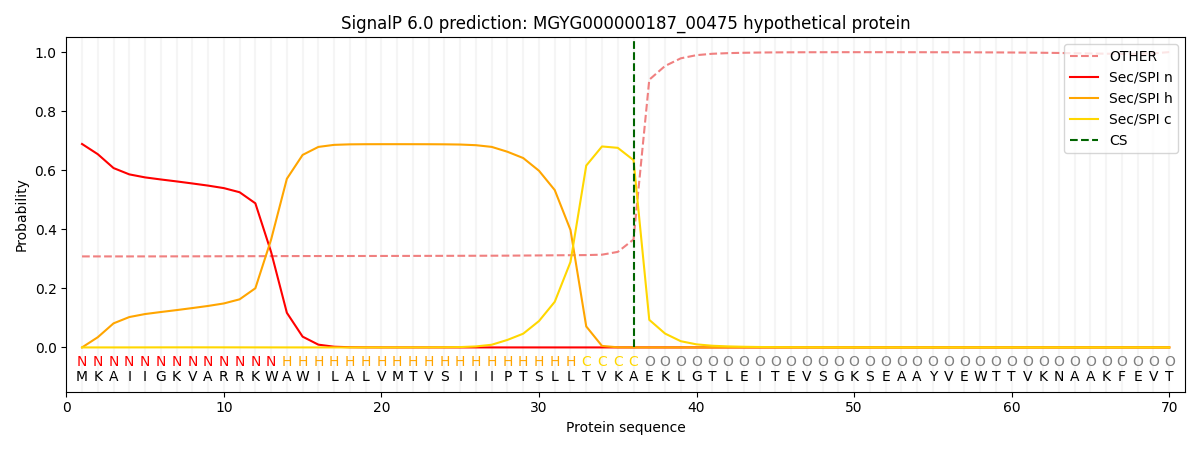
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.323781 | 0.667965 | 0.004226 | 0.002607 | 0.000725 | 0.000684 |