You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000187_00476
You are here: Home > Sequence: MGYG000000187_00476
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
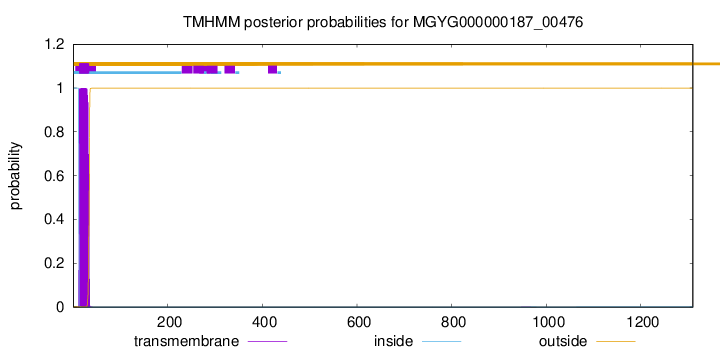
TMHMM annotations
Basic Information help
| Species | Lachnospira sp003537285 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Lachnospira; Lachnospira sp003537285 | |||||||||||
| CAZyme ID | MGYG000000187_00476 | |||||||||||
| CAZy Family | PL1 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 20179; End: 24111 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| PL9 | 652 | 1051 | 3.9e-111 | 0.9653333333333334 |
| PL1 | 229 | 414 | 6.6e-43 | 0.905940594059406 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3866 | PelB | 4.28e-53 | 102 | 533 | 2 | 344 | Pectate lyase [Carbohydrate transport and metabolism]. |
| smart00656 | Amb_all | 8.04e-32 | 250 | 416 | 17 | 190 | Amb_all domain. |
| pfam00544 | Pec_lyase_C | 4.07e-15 | 250 | 412 | 35 | 211 | Pectate lyase. This enzyme forms a right handed beta helix structure. Pectate lyase is an enzyme involved in the maceration and soft rotting of plant tissue. |
| cd14251 | PL-6 | 2.45e-07 | 666 | 807 | 1 | 134 | Polysaccharide Lyase Family 6. Polysaccharide Lyase Family 6 is a family of beta-helical polysaccharide lyases. Members include alginate lyase (EC 4.2.2.3) and chondroitinase B (EC 4.2.2.19). Chondroitinase B is an enzyme that only cleaves the beta-(1,4)-linkage of dermatan sulfate (DS), leading to 4,5-unsaturated dermatan sulfate disaccharides as the product. DS is a highly sulfated, unbranched polysaccharide belonging to a family of glycosaminoglycans (GAGs) composed of alternating hexosamine (gluco- or galactosamine) and uronic acid (D-glucuronic or L-iduronic acid) moieties. DS contains alternating 1,4-beta-D-galactosamine (GalNac) and 1,3-alpha-L-iduronic acid units. The related chondroitin sulfate (CS) contains alternating GalNac and 1,3-beta-D-glucuronic acid units. Alginate lyases (known as either mannuronate (EC 4.2.2.3) or guluronate lyases (EC 4.2.2.11) catalyze the degradation of alginate, a copolymer of alpha-L-guluronate and its C5 epimer beta-D-mannuronate. |
| pfam14592 | Chondroitinas_B | 1.58e-06 | 666 | 806 | 1 | 134 | Chondroitinase B. This family includes chondroitinases. These enzymes cleave the glycosaminoglycan dermatan sulfate. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ACR72247.1 | 0.0 | 1 | 1310 | 1 | 1286 |
| ACR72246.1 | 0.0 | 1 | 1309 | 1 | 1472 |
| ABX41986.1 | 3.63e-259 | 6 | 1049 | 2 | 1144 |
| QMW93302.1 | 1.32e-256 | 27 | 1047 | 41 | 1072 |
| BBK78741.1 | 1.32e-256 | 27 | 1047 | 41 | 1072 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1RU4_A | 7.08e-32 | 675 | 939 | 41 | 293 | ChainA, Pectate lyase [Dickeya chrysanthemi] |
| 3VMV_A | 2.17e-24 | 186 | 414 | 18 | 248 | Crystalstructure of pectate lyase Bsp165PelA from Bacillus sp. N165 [Bacillus sp. N16-5],3VMW_A Crystal structure of pectate lyase Bsp165PelA from Bacillus sp. N165 in complex with trigalacturonate [Bacillus sp. N16-5] |
| 1VBL_A | 1.85e-18 | 246 | 412 | 129 | 330 | Structureof the thermostable pectate lyase PL 47 [Bacillus sp. TS-47] |
| 5OLQ_A | 3.44e-16 | 760 | 940 | 140 | 321 | Rhamnogalacturonanlyase [Bacteroides thetaiotaomicron],5OLQ_B Rhamnogalacturonan lyase [Bacteroides thetaiotaomicron],5OLQ_C Rhamnogalacturonan lyase [Bacteroides thetaiotaomicron],5OLR_A Rhamnogalacturonan lyase [Bacteroides thetaiotaomicron],5OLR_B Rhamnogalacturonan lyase [Bacteroides thetaiotaomicron],5OLR_C Rhamnogalacturonan lyase [Bacteroides thetaiotaomicron],5OLS_A Rhamnogalacturonan lyase [Bacteroides thetaiotaomicron] |
| 1AIR_A | 4.45e-15 | 151 | 438 | 2 | 282 | ChainA, PECTATE LYASE C [Dickeya chrysanthemi],1O88_A Chain A, Pectate Lyase C [Dickeya chrysanthemi],1O8D_A Chain A, PECTATE LYASE C [Dickeya chrysanthemi],1O8E_A Chain A, PECTATE LYASE C [Dickeya chrysanthemi],1O8F_A Chain A, PECTATE LYASE C [Dickeya chrysanthemi],1O8G_A Chain A, PECTATE LYASE C [Dickeya chrysanthemi],1O8H_A Chain A, PECTATE LYASE C [Dickeya chrysanthemi],1O8I_A Chain A, PECTATE LYASE C [Dickeya chrysanthemi],1O8J_A Chain A, PECTATE LYASE C [Dickeya chrysanthemi],1O8K_A Chain A, PECTATE LYASE C [Dickeya chrysanthemi],1O8L_A Chain A, PECTATE LYASE C [Dickeya chrysanthemi],1O8M_A Chain A, PECTATE LYASE C [Dickeya chrysanthemi],1PLU_A Chain A, Protein (pectate Lyase C) [Dickeya chrysanthemi],2PEC_A Chain A, PECTATE LYASE C [Dickeya chrysanthemi] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P0C1A6 | 5.52e-32 | 675 | 939 | 66 | 318 | Pectate lyase L OS=Dickeya chrysanthemi OX=556 GN=pelL PE=3 SV=1 |
| P0C1A7 | 6.00e-31 | 675 | 939 | 66 | 318 | Pectate lyase L OS=Dickeya dadantii (strain 3937) OX=198628 GN=pelL PE=1 SV=1 |
| P22751 | 2.56e-27 | 737 | 941 | 460 | 642 | Pectate disaccharide-lyase OS=Dickeya chrysanthemi OX=556 GN=pelX PE=1 SV=1 |
| Q8GCB2 | 9.39e-23 | 193 | 421 | 67 | 281 | Pectate trisaccharide-lyase OS=Bacillus licheniformis OX=1402 GN=pelA PE=1 SV=1 |
| Q65DC2 | 9.39e-23 | 193 | 421 | 67 | 281 | Pectate trisaccharide-lyase OS=Bacillus licheniformis (strain ATCC 14580 / DSM 13 / JCM 2505 / CCUG 7422 / NBRC 12200 / NCIMB 9375 / NCTC 10341 / NRRL NRS-1264 / Gibson 46) OX=279010 GN=BLi04129 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
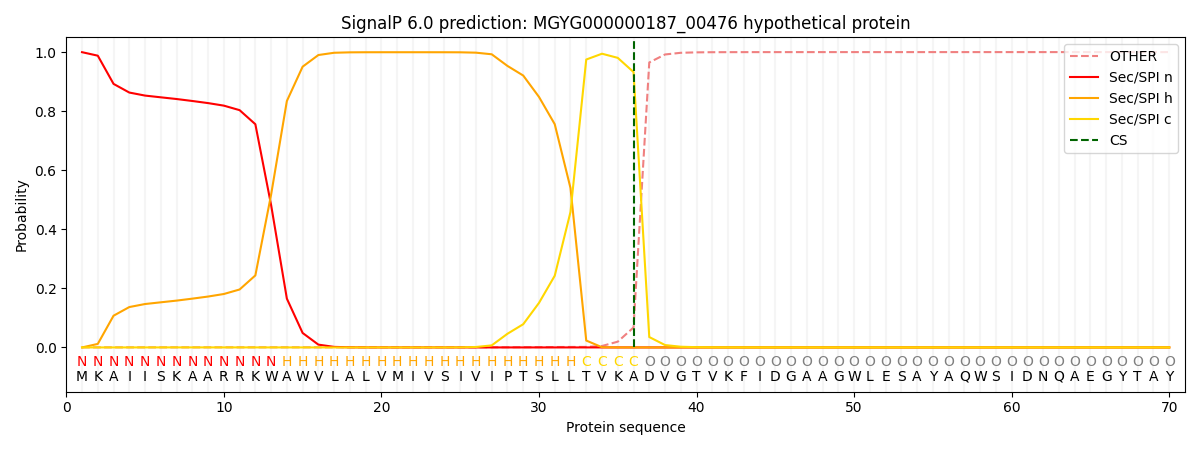
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000773 | 0.998479 | 0.000222 | 0.000191 | 0.000160 | 0.000163 |