You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000193_00672
You are here: Home > Sequence: MGYG000000193_00672
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | KLE1615 sp900066985 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; KLE1615; KLE1615 sp900066985 | |||||||||||
| CAZyme ID | MGYG000000193_00672 | |||||||||||
| CAZy Family | GH28 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 64894; End: 67806 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH28 | 51 | 369 | 1e-36 | 0.7476923076923077 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG5434 | Pgu1 | 3.42e-38 | 5 | 392 | 65 | 429 | Polygalacturonase [Carbohydrate transport and metabolism]. |
| pfam00583 | Acetyltransf_1 | 1.65e-07 | 824 | 944 | 10 | 116 | Acetyltransferase (GNAT) family. This family contains proteins with N-acetyltransferase functions such as Elp3-related proteins. |
| COG0456 | RimI | 5.29e-05 | 819 | 948 | 27 | 153 | Ribosomal protein S18 acetylase RimI and related acetyltransferases [Translation, ribosomal structure and biogenesis]. |
| cd04301 | NAT_SF | 8.13e-04 | 849 | 930 | 1 | 65 | N-Acyltransferase superfamily: Various enzymes that characteristically catalyze the transfer of an acyl group to a substrate. NAT (N-Acyltransferase) is a large superfamily of enzymes that mostly catalyze the transfer of an acyl group to a substrate and are implicated in a variety of functions, ranging from bacterial antibiotic resistance to circadian rhythms in mammals. Members include GCN5-related N-Acetyltransferases (GNAT) such as Aminoglycoside N-acetyltransferases, Histone N-acetyltransferase (HAT) enzymes, and Serotonin N-acetyltransferase, which catalyze the transfer of an acetyl group to a substrate. The kinetic mechanism of most GNATs involves the ordered formation of a ternary complex: the reaction begins with Acetyl Coenzyme A (AcCoA) binding, followed by binding of substrate, then direct transfer of the acetyl group from AcCoA to the substrate, followed by product and subsequent CoA release. Other family members include Arginine/ornithine N-succinyltransferase, Myristoyl-CoA: protein N-myristoyltransferase, and Acyl-homoserinelactone synthase which have a similar catalytic mechanism but differ in types of acyl groups transferred. Leucyl/phenylalanyl-tRNA-protein transferase and FemXAB nonribosomal peptidyltransferases which catalyze similar peptidyltransferase reactions are also included. |
| pfam13508 | Acetyltransf_7 | 0.001 | 845 | 944 | 1 | 81 | Acetyltransferase (GNAT) domain. This domain catalyzes N-acetyltransferase reactions. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BCJ93914.1 | 1.09e-312 | 1 | 842 | 1 | 865 |
| CBL02329.1 | 9.06e-276 | 4 | 760 | 45 | 801 |
| AXB28928.1 | 1.66e-275 | 4 | 760 | 63 | 819 |
| AHG93184.1 | 2.85e-96 | 39 | 655 | 50 | 492 |
| QOY86475.1 | 3.89e-94 | 27 | 655 | 28 | 477 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3JUR_A | 1.19e-10 | 27 | 278 | 32 | 287 | Thecrystal structure of a hyperthermoactive Exopolygalacturonase from Thermotoga maritima [Thermotoga maritima],3JUR_B The crystal structure of a hyperthermoactive Exopolygalacturonase from Thermotoga maritima [Thermotoga maritima],3JUR_C The crystal structure of a hyperthermoactive Exopolygalacturonase from Thermotoga maritima [Thermotoga maritima],3JUR_D The crystal structure of a hyperthermoactive Exopolygalacturonase from Thermotoga maritima [Thermotoga maritima] |
| 4MXN_A | 2.33e-08 | 39 | 250 | 37 | 215 | Crystalstructure of a putative glycosyl hydrolase (PARMER_00599) from Parabacteroides merdae ATCC 43184 at 1.95 A resolution [Parabacteroides merdae ATCC 43184],4MXN_B Crystal structure of a putative glycosyl hydrolase (PARMER_00599) from Parabacteroides merdae ATCC 43184 at 1.95 A resolution [Parabacteroides merdae ATCC 43184],4MXN_C Crystal structure of a putative glycosyl hydrolase (PARMER_00599) from Parabacteroides merdae ATCC 43184 at 1.95 A resolution [Parabacteroides merdae ATCC 43184],4MXN_D Crystal structure of a putative glycosyl hydrolase (PARMER_00599) from Parabacteroides merdae ATCC 43184 at 1.95 A resolution [Parabacteroides merdae ATCC 43184] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as OTHER
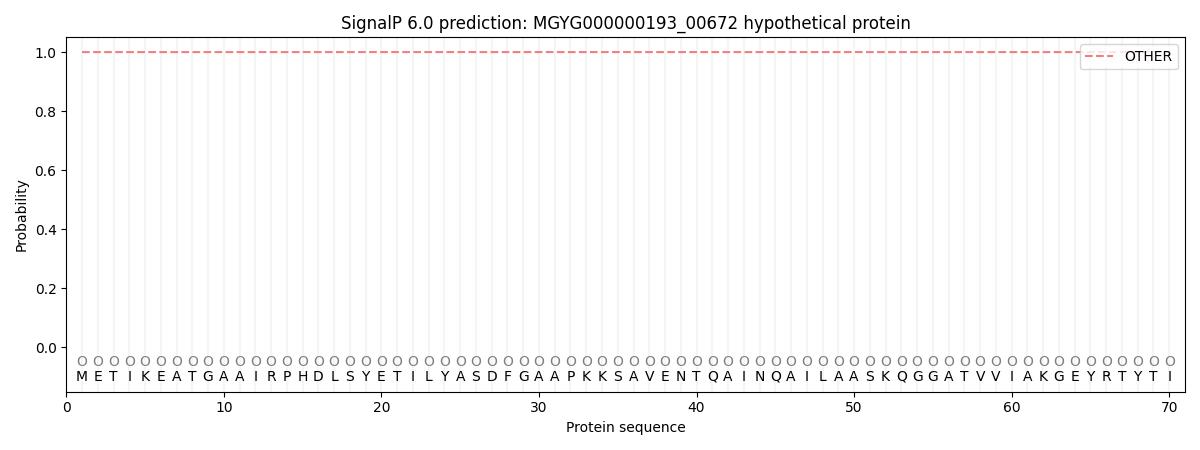
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000050 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |
