You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000196_00136
You are here: Home > Sequence: MGYG000000196_00136
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Bacteroides thetaiotaomicron | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; Bacteroides thetaiotaomicron | |||||||||||
| CAZyme ID | MGYG000000196_00136 | |||||||||||
| CAZy Family | GH13 | |||||||||||
| CAZyme Description | Alpha-amylase SusG | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 170994; End: 173072 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH13 | 80 | 540 | 1.2e-136 | 0.9968152866242038 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd11316 | AmyAc_bac2_AmyA | 8.91e-164 | 61 | 607 | 1 | 403 | Alpha amylase catalytic domain found in bacterial Alpha-amylases (also called 1,4-alpha-D-glucan-4-glucanohydrolase). AmyA (EC 3.2.1.1) catalyzes the hydrolysis of alpha-(1,4) glycosidic linkages of glycogen, starch, related polysaccharides, and some oligosaccharides. This group includes Chloroflexi, Dictyoglomi, and Fusobacteria. The Alpha-amylase family comprises the largest family of glycoside hydrolases (GH), with the majority of enzymes acting on starch, glycogen, and related oligo- and polysaccharides. These proteins catalyze the transformation of alpha-1,4 and alpha-1,6 glucosidic linkages with retention of the anomeric center. The protein is described as having 3 domains: A, B, C. A is a (beta/alpha) 8-barrel; B is a loop between the beta 3 strand and alpha 3 helix of A; C is the C-terminal extension characterized by a Greek key. The majority of the enzymes have an active site cleft found between domains A and B where a triad of catalytic residues (Asp, Glu and Asp) performs catalysis. Other members of this family have lost the catalytic activity as in the case of the human 4F2hc, or only have 2 residues that serve as the catalytic nucleophile and the acid/base, such as Thermus A4 beta-galactosidase with 2 Glu residues (GH42) and human alpha-galactosidase with 2 Asp residues (GH31). The family members are quite extensive and include: alpha amylase, maltosyltransferase, cyclodextrin glycotransferase, maltogenic amylase, neopullulanase, isoamylase, 1,4-alpha-D-glucan maltotetrahydrolase, 4-alpha-glucotransferase, oligo-1,6-glucosidase, amylosucrase, sucrose phosphorylase, and amylomaltase. |
| cd11333 | AmyAc_SI_OligoGlu_DGase | 3.50e-68 | 64 | 599 | 6 | 427 | Alpha amylase catalytic domain found in Sucrose isomerases, oligo-1,6-glucosidase (also called isomaltase; sucrase-isomaltase; alpha-limit dextrinase), dextran glucosidase (also called glucan 1,6-alpha-glucosidase), and related proteins. The sucrose isomerases (SIs) Isomaltulose synthase (EC 5.4.99.11) and Trehalose synthase (EC 5.4.99.16) catalyze the isomerization of sucrose and maltose to produce isomaltulose and trehalulose, respectively. Oligo-1,6-glucosidase (EC 3.2.1.10) hydrolyzes the alpha-1,6-glucosidic linkage of isomaltooligosaccharides, pannose, and dextran. Unlike alpha-1,4-glucosidases (EC 3.2.1.20), it fails to hydrolyze the alpha-1,4-glucosidic bonds of maltosaccharides. Dextran glucosidase (DGase, EC 3.2.1.70) hydrolyzes alpha-1,6-glucosidic linkages at the non-reducing end of panose, isomaltooligosaccharides and dextran to produce alpha-glucose.The common reaction chemistry of the alpha-amylase family enzymes is based on a two-step acid catalytic mechanism that requires two critical carboxylates: one acting as a general acid/base (Glu) and the other as a nucleophile (Asp). Both hydrolysis and transglycosylation proceed via the nucleophilic substitution reaction between the anomeric carbon, C1 and a nucleophile. Both enzymes contain the three catalytic residues (Asp, Glu and Asp) common to the alpha-amylase family as well as two histidine residues which are predicted to be critical to binding the glucose residue adjacent to the scissile bond in the substrates. The Alpha-amylase family comprises the largest family of glycoside hydrolases (GH), with the majority of enzymes acting on starch, glycogen, and related oligo- and polysaccharides. These proteins catalyze the transformation of alpha-1,4 and alpha-1,6 glucosidic linkages with retention of the anomeric center. The protein is described as having 3 domains: A, B, C. A is a (beta/alpha) 8-barrel; B is a loop between the beta 3 strand and alpha 3 helix of A; C is the C-terminal extension characterized by a Greek key. The majority of the enzymes have an active site cleft found between domains A and B where a triad of catalytic residues performs catalysis. Other members of this family have lost the catalytic activity as in the case of the human 4F2hc, or only have 2 residues that serve as the catalytic nucleophile and the acid/base, such as Thermus A4 beta-galactosidase with 2 Glu residues (GH42) and human alpha-galactosidase with 2 Asp residues (GH31). The family members are quite extensive and include: alpha amylase, maltosyltransferase, cyclodextrin glycotransferase, maltogenic amylase, neopullulanase, isoamylase, 1,4-alpha-D-glucan maltotetrahydrolase, 4-alpha-glucotransferase, oligo-1,6-glucosidase, amylosucrase, sucrose phosphorylase, and amylomaltase. |
| cd12961 | CBM58_SusG | 4.99e-63 | 222 | 331 | 1 | 110 | Carbohydrate-binding module 58 from Bacteroides thetaiotaomicron SusG and similar CBMs. This group includes the starch-specific CBM (carbohydrate-binding module) of SusG, a cell surface lipoprotein within the Sus (Starch-utilization system) system of the Human gut symbiont Bacteroides thetaiotaomicron. It represents the CBM58 class of CBMs in the carbohydrate active enzymes (CAZy) database. SusG is an alpha-amylase, and is essential for growth on high molecular weight starch. SusG-CBM58 binds maltooligosaccharide distal to, and on the opposite side of, the amylase catalytic site; it is one of two starch-binding sites in SusG, the other being adjacent to the active site. SusG-CBM58 is required for efficient degradation of insoluble starch by the purified enzyme. Its starch-binding site contains an arc of aromatic amino acids for hydrophobic stacking with glucose, and hydrogen-bonding acceptors and donors for interacting with the O-2 and O-3 of glucose. It may play a role in product exchange with other Sus components. |
| cd11331 | AmyAc_OligoGlu_like | 2.16e-59 | 62 | 607 | 7 | 449 | Alpha amylase catalytic domain found in oligo-1,6-glucosidase (also called isomaltase; sucrase-isomaltase; alpha-limit dextrinase) and related proteins. Oligo-1,6-glucosidase (EC 3.2.1.10) hydrolyzes the alpha-1,6-glucosidic linkage of isomalto-oligosaccharides, pannose, and dextran. Unlike alpha-1,4-glucosidases (EC 3.2.1.20), it fails to hydrolyze the alpha-1,4-glucosidic bonds of maltosaccharides. The Alpha-amylase family comprises the largest family of glycoside hydrolases (GH), with the majority of enzymes acting on starch, glycogen, and related oligo- and polysaccharides. These proteins catalyze the transformation of alpha-1,4 and alpha-1,6 glucosidic linkages with retention of the anomeric center. The protein is described as having 3 domains: A, B, C. A is a (beta/alpha) 8-barrel; B is a loop between the beta 3 strand and alpha 3 helix of A; C is the C-terminal extension characterized by a Greek key. The majority of the enzymes have an active site cleft found between domains A and B where a triad of catalytic residues (Asp, Glu and Asp) performs catalysis. Other members of this family have lost the catalytic activity as in the case of the human 4F2hc, or only have 2 residues that serve as the catalytic nucleophile and the acid/base, such as Thermus A4 beta-galactosidase with 2 Glu residues (GH42) and human alpha-galactosidase with 2 Asp residues (GH31). The family members are quite extensive and include: alpha amylase, maltosyltransferase, cyclodextrin glycotransferase, maltogenic amylase, neopullulanase, isoamylase, 1,4-alpha-D-glucan maltotetrahydrolase, 4-alpha-glucotransferase, oligo-1,6-glucosidase, amylosucrase, sucrose phosphorylase, and amylomaltase. |
| pfam00128 | Alpha-amylase | 1.62e-58 | 80 | 542 | 1 | 333 | Alpha amylase, catalytic domain. Alpha amylase is classified as family 13 of the glycosyl hydrolases. The structure is an 8 stranded alpha/beta barrel containing the active site, interrupted by a ~70 a.a. calcium-binding domain protruding between beta strand 3 and alpha helix 3, and a carboxyl-terminal Greek key beta-barrel domain. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QQA08186.1 | 0.0 | 1 | 692 | 1 | 692 |
| ALJ44114.1 | 0.0 | 1 | 692 | 1 | 692 |
| QMW86069.1 | 0.0 | 1 | 692 | 1 | 692 |
| AAO78803.1 | 0.0 | 1 | 692 | 1 | 692 |
| CAZ78746.1 | 0.0 | 1 | 692 | 1 | 692 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6BS6_A | 0.0 | 24 | 692 | 2 | 670 | SusGwith mixed linkage amylosaccharide [Bacteroides thetaiotaomicron VPI-5482],6BS6_B SusG with mixed linkage amylosaccharide [Bacteroides thetaiotaomicron VPI-5482] |
| 3K8L_A | 0.0 | 24 | 692 | 1 | 669 | ChainA, Alpha-amylase, susG [Bacteroides thetaiotaomicron],3K8L_B Chain B, Alpha-amylase, susG [Bacteroides thetaiotaomicron] |
| 3K8K_A | 0.0 | 24 | 692 | 1 | 669 | Crystalstructure of SusG [Bacteroides thetaiotaomicron],3K8K_B Crystal structure of SusG [Bacteroides thetaiotaomicron],3K8M_A Crystal structure of SusG with acarbose [Bacteroides thetaiotaomicron],3K8M_B Crystal structure of SusG with acarbose [Bacteroides thetaiotaomicron] |
| 1WZA_A | 6.93e-57 | 64 | 689 | 8 | 486 | Crystalstructure of alpha-amylase from H.orenii [Halothermothrix orenii] |
| 7JJT_A | 2.71e-55 | 39 | 681 | 16 | 508 | ChainA, Alpha-amylase [Ruminococcus bromii] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q8A1G3 | 0.0 | 1 | 692 | 1 | 692 | Alpha-amylase SusG OS=Bacteroides thetaiotaomicron (strain ATCC 29148 / DSM 2079 / JCM 5827 / CCUG 10774 / NCTC 10582 / VPI-5482 / E50) OX=226186 GN=susG PE=1 SV=1 |
| P14899 | 2.76e-69 | 62 | 690 | 36 | 498 | Alpha-amylase 3 OS=Dictyoglomus thermophilum (strain ATCC 35947 / DSM 3960 / H-6-12) OX=309799 GN=amyC PE=3 SV=2 |
| P20845 | 2.27e-59 | 62 | 656 | 41 | 491 | Alpha-amylase OS=Priestia megaterium OX=1404 PE=1 SV=1 |
| P39795 | 4.28e-35 | 62 | 690 | 13 | 557 | Trehalose-6-phosphate hydrolase OS=Bacillus subtilis (strain 168) OX=224308 GN=treA PE=1 SV=2 |
| Q45101 | 1.54e-32 | 62 | 666 | 9 | 535 | Oligo-1,6-glucosidase OS=Weizmannia coagulans OX=1398 GN=malL PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
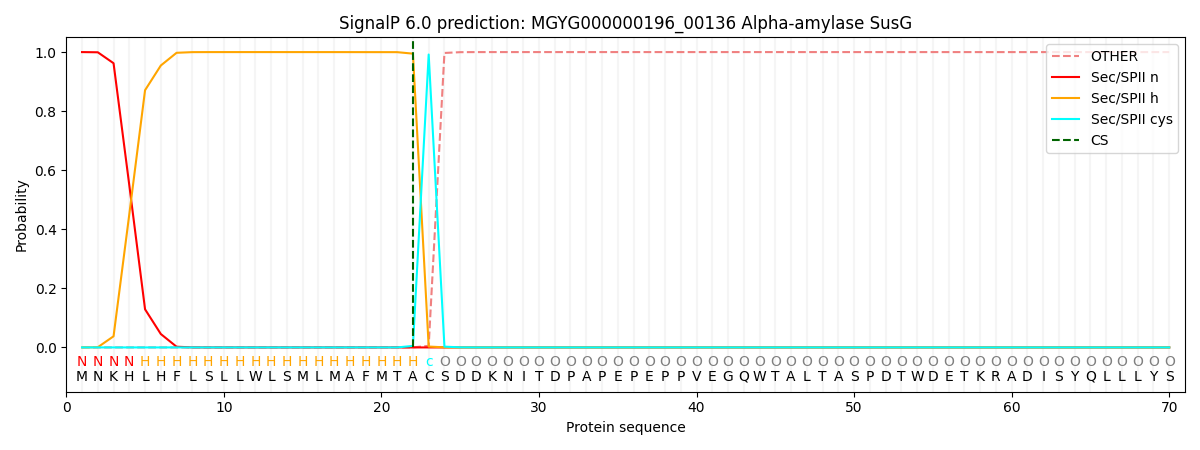
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000002 | 1.000054 | 0.000000 | 0.000000 | 0.000000 |
