You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000199_00124
You are here: Home > Sequence: MGYG000000199_00124
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Streptococcus parasanguinis_B | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Lactobacillales; Streptococcaceae; Streptococcus; Streptococcus parasanguinis_B | |||||||||||
| CAZyme ID | MGYG000000199_00124 | |||||||||||
| CAZy Family | GH25 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 132829; End: 135282 Strand: + | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam08481 | GBS_Bsp-like | 1.51e-33 | 445 | 533 | 1 | 89 | GBS Bsp-like repeat. This domain is found as a repeat in a number of Streptococcus proteins including some hypothetical proteins and Bsp. Bsp is a protein of group B Streptococcus (GBS) which might control cell morphology. |
| pfam08481 | GBS_Bsp-like | 5.48e-19 | 241 | 326 | 3 | 88 | GBS Bsp-like repeat. This domain is found as a repeat in a number of Streptococcus proteins including some hypothetical proteins and Bsp. Bsp is a protein of group B Streptococcus (GBS) which might control cell morphology. |
| pfam08481 | GBS_Bsp-like | 1.09e-18 | 343 | 428 | 3 | 88 | GBS Bsp-like repeat. This domain is found as a repeat in a number of Streptococcus proteins including some hypothetical proteins and Bsp. Bsp is a protein of group B Streptococcus (GBS) which might control cell morphology. |
| pfam08481 | GBS_Bsp-like | 1.98e-18 | 140 | 224 | 4 | 88 | GBS Bsp-like repeat. This domain is found as a repeat in a number of Streptococcus proteins including some hypothetical proteins and Bsp. Bsp is a protein of group B Streptococcus (GBS) which might control cell morphology. |
| pfam05257 | CHAP | 1.31e-08 | 746 | 791 | 42 | 83 | CHAP domain. This domain corresponds to an amidase function. Many of these proteins are involved in cell wall metabolism of bacteria. This domain is found at the N-terminus of Escherichia coli gss, where it functions as a glutathionylspermidine amidase EC:3.5.1.78. This domain is found to be the catalytic domain of PlyCA. CHAP is the amidase domain of bifunctional Escherichia coli glutathionylspermidine synthetase/amidase, and it catalyzes the hydrolysis of Gsp (glutathionylspermidine) into glutathione and spermidine. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QWL83937.1 | 0.0 | 131 | 817 | 462 | 1156 |
| AFJ26940.1 | 0.0 | 131 | 817 | 680 | 1374 |
| AEH56405.1 | 0.0 | 131 | 817 | 574 | 1268 |
| AGY39024.1 | 0.0 | 127 | 643 | 459 | 979 |
| AAN64579.1 | 1.27e-274 | 145 | 817 | 703 | 1366 |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
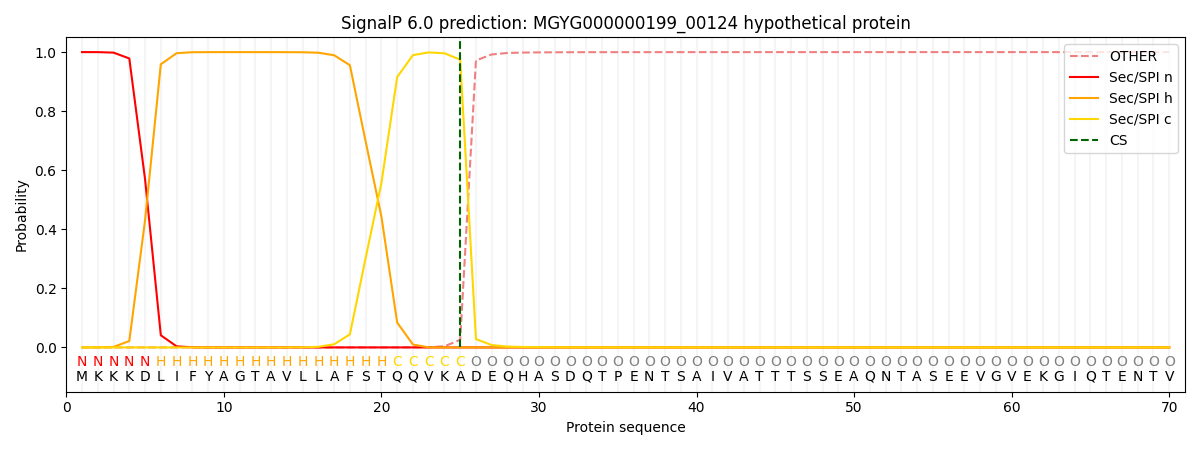
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000392 | 0.998740 | 0.000363 | 0.000167 | 0.000164 | 0.000149 |
