You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000202_00260
You are here: Home > Sequence: MGYG000000202_00260
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
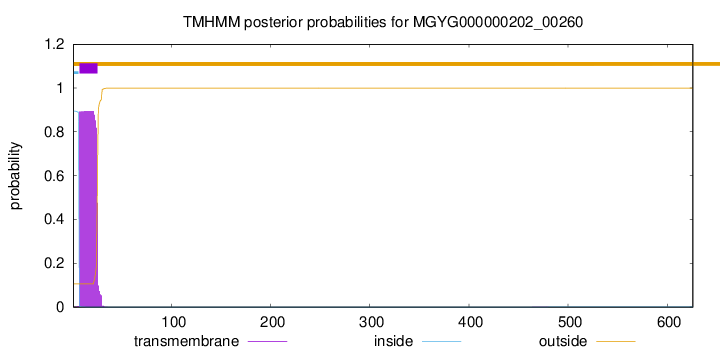
TMHMM annotations
Basic Information help
| Species | AF33-28 sp003477885 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; AF33-28; AF33-28 sp003477885 | |||||||||||
| CAZyme ID | MGYG000000202_00260 | |||||||||||
| CAZy Family | GH30 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 315721; End: 317601 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH30 | 31 | 468 | 3.4e-136 | 0.9978070175438597 |
| CBM13 | 528 | 624 | 9.6e-18 | 0.5 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam14587 | Glyco_hydr_30_2 | 5.09e-29 | 30 | 341 | 3 | 356 | O-Glycosyl hydrolase family 30. |
| COG5520 | XynC | 5.31e-22 | 14 | 471 | 18 | 429 | O-Glycosyl hydrolase [Cell wall/membrane/envelope biogenesis]. |
| pfam14200 | RicinB_lectin_2 | 1.94e-21 | 523 | 610 | 2 | 89 | Ricin-type beta-trefoil lectin domain-like. |
| pfam14200 | RicinB_lectin_2 | 7.15e-14 | 569 | 623 | 1 | 55 | Ricin-type beta-trefoil lectin domain-like. |
| cd00161 | RICIN | 9.10e-12 | 487 | 622 | 1 | 124 | Ricin-type beta-trefoil; Carbohydrate-binding domain formed from presumed gene triplication. The domain is found in a variety of molecules serving diverse functions such as enzymatic activity, inhibitory toxicity and signal transduction. Highly specific ligand binding occurs on exposed surfaces of the compact domain sturcture. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QLG42765.1 | 5.50e-132 | 31 | 477 | 19 | 466 |
| QZN75685.1 | 8.40e-131 | 31 | 474 | 50 | 494 |
| APO48420.1 | 8.73e-131 | 31 | 474 | 19 | 463 |
| AWB44606.1 | 2.06e-129 | 28 | 473 | 41 | 487 |
| QOS82337.1 | 7.80e-129 | 31 | 477 | 19 | 466 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3CLW_A | 9.69e-16 | 33 | 382 | 10 | 406 | Crystalstructure of conserved exported protein from Bacteroides fragilis [Bacteroides fragilis NCTC 9343],3CLW_B Crystal structure of conserved exported protein from Bacteroides fragilis [Bacteroides fragilis NCTC 9343],3CLW_C Crystal structure of conserved exported protein from Bacteroides fragilis [Bacteroides fragilis NCTC 9343],3CLW_D Crystal structure of conserved exported protein from Bacteroides fragilis [Bacteroides fragilis NCTC 9343],3CLW_E Crystal structure of conserved exported protein from Bacteroides fragilis [Bacteroides fragilis NCTC 9343],3CLW_F Crystal structure of conserved exported protein from Bacteroides fragilis [Bacteroides fragilis NCTC 9343] |
| 4CKQ_A | 1.04e-13 | 130 | 469 | 92 | 409 | ChainA, Carbohydrate Binding Family 6 [Acetivibrio thermocellus],4UQ9_A Chain A, Carbohydrate Binding Family 6 [Acetivibrio thermocellus],4UQB_A Chain A, Carbohydrate Binding Family 6 [Acetivibrio thermocellus],4UQC_A Chain A, Carbohydrate Binding Family 6 [Acetivibrio thermocellus],4UQD_A Chain A, Carbohydrate Binding Family 6 [Acetivibrio thermocellus],4UQE_A Chain A, Carbohydrate Binding Family 6 [Acetivibrio thermocellus] |
| 4UQA_A | 2.46e-13 | 130 | 469 | 92 | 409 | ChainA, Carbohydrate Binding Family 6 [Acetivibrio thermocellus] |
| 5A6L_A | 4.36e-13 | 130 | 469 | 92 | 409 | ChainA, Carbohydrate Binding Family 6 [Acetivibrio thermocellus],5A6M_A Chain A, Carbohydrate Binding Family 6 [Acetivibrio thermocellus ATCC 27405] |
| 4QAW_A | 9.36e-13 | 130 | 471 | 72 | 389 | Structureof modular Xyn30D from Paenibacillus barcinonensis [Paenibacillus barcinonensis],4QAW_B Structure of modular Xyn30D from Paenibacillus barcinonensis [Paenibacillus barcinonensis],4QAW_C Structure of modular Xyn30D from Paenibacillus barcinonensis [Paenibacillus barcinonensis],4QAW_D Structure of modular Xyn30D from Paenibacillus barcinonensis [Paenibacillus barcinonensis],4QAW_E Structure of modular Xyn30D from Paenibacillus barcinonensis [Paenibacillus barcinonensis],4QAW_F Structure of modular Xyn30D from Paenibacillus barcinonensis [Paenibacillus barcinonensis],4QAW_G Structure of modular Xyn30D from Paenibacillus barcinonensis [Paenibacillus barcinonensis],4QAW_H Structure of modular Xyn30D from Paenibacillus barcinonensis [Paenibacillus barcinonensis] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q76FP5 | 1.53e-47 | 11 | 473 | 4 | 476 | Endo-beta-1,6-galactanase OS=Hypocrea rufa OX=5547 GN=6GAL PE=1 SV=1 |
| A0A401ETL2 | 8.49e-25 | 30 | 528 | 38 | 695 | Exo-beta-1,6-galactobiohydrolase OS=Bifidobacterium longum subsp. longum (strain ATCC 15707 / DSM 20219 / JCM 1217 / NCTC 11818 / E194b) OX=565042 GN=bl1,6Gal PE=1 SV=1 |
| Q45070 | 1.35e-10 | 158 | 471 | 127 | 421 | Glucuronoxylanase XynC OS=Bacillus subtilis (strain 168) OX=224308 GN=xynC PE=1 SV=1 |
| Q6YK37 | 1.30e-09 | 130 | 470 | 105 | 421 | Glucuronoxylanase XynC OS=Bacillus subtilis OX=1423 GN=xynC PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
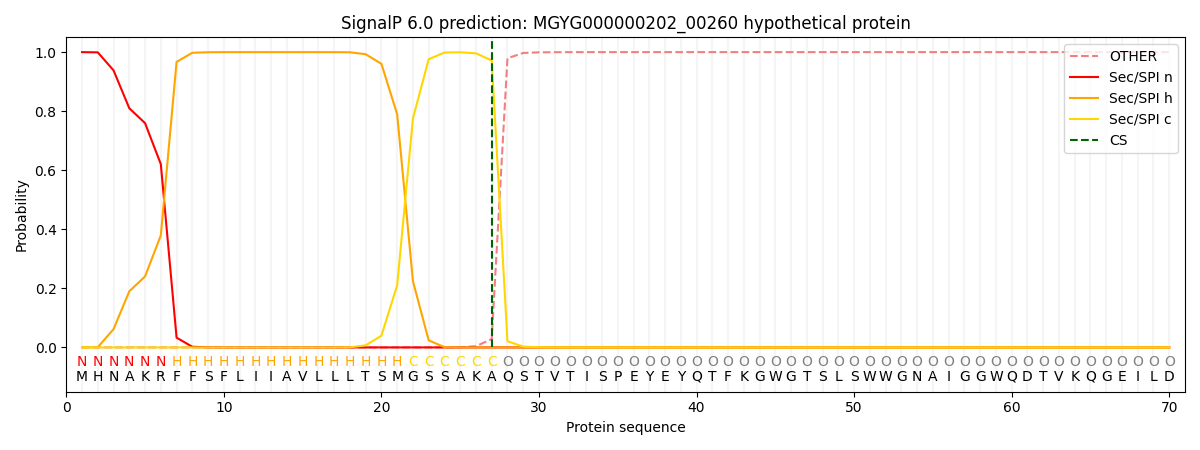
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000282 | 0.998293 | 0.000822 | 0.000219 | 0.000204 | 0.000165 |