You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000202_02000
You are here: Home > Sequence: MGYG000000202_02000
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
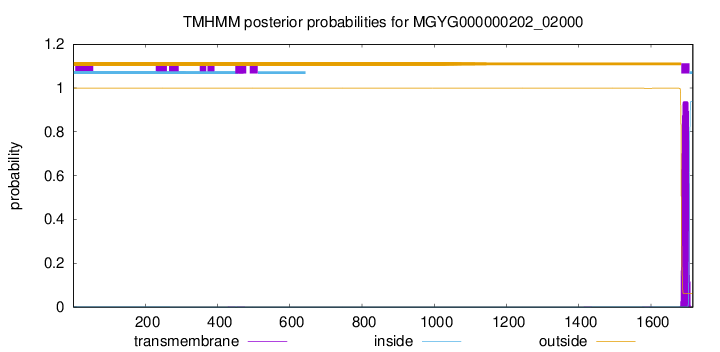
TMHMM annotations
Basic Information help
| Species | AF33-28 sp003477885 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; AF33-28; AF33-28 sp003477885 | |||||||||||
| CAZyme ID | MGYG000000202_02000 | |||||||||||
| CAZy Family | GH123 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 81500; End: 86650 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM66 | 1164 | 1318 | 9.3e-20 | 0.9354838709677419 |
| GH123 | 253 | 543 | 1.6e-18 | 0.4944237918215613 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam06439 | DUF1080 | 6.91e-10 | 1156 | 1321 | 7 | 181 | Domain of Unknown Function (DUF1080). This family has structural similarity to an endo-1,3-1,4-beta glucanase belonging to glycoside hydrolase family 16. However, the structure surrounding the active site differs from that of the endo-1,3-1,4-beta glucanase. |
| pfam13320 | DUF4091 | 1.42e-08 | 464 | 525 | 1 | 66 | Domain of unknown function (DUF4091). This presumed domain is functionally uncharacterized. This domain family is found in bacteria, archaea and eukaryotes, and is approximately 70 amino acids in length. There is a single completely conserved residue G that may be functionally important. |
| TIGR03804 | para_beta_helix | 0.010 | 1508 | 1540 | 5 | 44 | parallel beta-helix repeat (two copies). This model represents a tandem pair of an approximately 22-amino acid (each) repeat homologous to the beta-strand repeats that stack in a right-handed parallel beta-helix in the periplasmic C-5 mannuronan epimerase, AlgA, of Pseudomonas aeruginosa. A homology domain consisting of a longer tandem array of these repeats is described in the SMART database as CASH (SM00722), and is found in many carbohydrate-binding proteins and sugar hydrolases. A single repeat is represented by SM00710. This TIGRFAMs model represents a flavor of the parallel beta-helix-forming repeat based on prokaryotic sequences only in its seed alignment, although it also finds many eukaryotic sequences. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QNE29218.1 | 2.27e-153 | 11 | 827 | 30 | 838 |
| QEV44207.1 | 5.58e-147 | 11 | 827 | 9 | 817 |
| QJD85658.1 | 2.87e-141 | 14 | 936 | 26 | 934 |
| QTH41935.1 | 6.27e-74 | 11 | 588 | 23 | 755 |
| BBI36344.1 | 1.53e-62 | 24 | 580 | 44 | 767 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5L7V_A | 2.03e-11 | 44 | 527 | 49 | 519 | ChainA, glycoside hydrolase [Phocaeicola vulgatus ATCC 8482],5L7V_B Chain B, glycoside hydrolase [Phocaeicola vulgatus ATCC 8482] |
| 5L7R_A | 2.09e-11 | 44 | 527 | 64 | 534 | ChainA, glycoside hydrolase [Phocaeicola vulgatus ATCC 8482],5L7R_B Chain B, glycoside hydrolase [Phocaeicola vulgatus ATCC 8482],5L7U_A Chain A, Glycoside hydrolase [Phocaeicola vulgatus ATCC 8482],5L7U_B Chain B, Glycoside hydrolase [Phocaeicola vulgatus ATCC 8482] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as OTHER
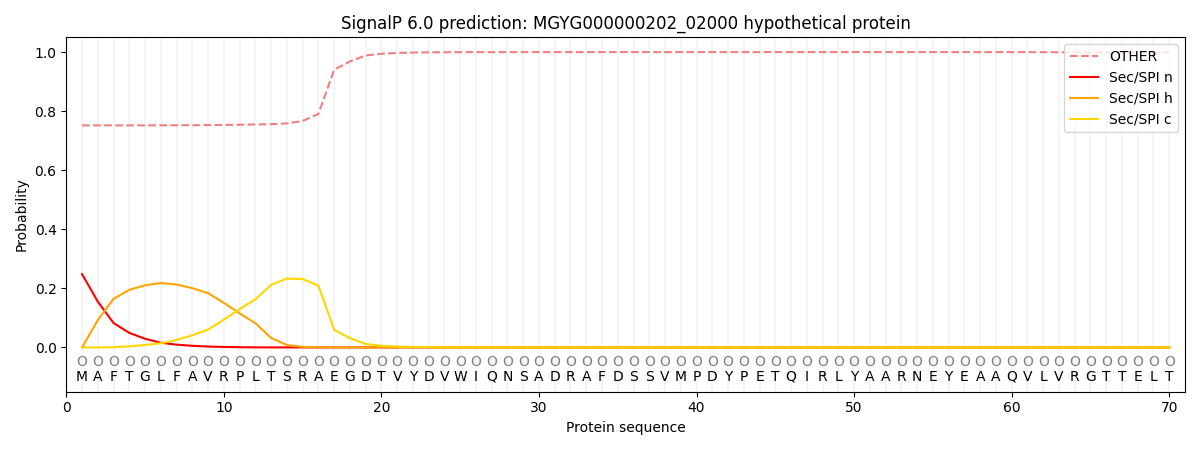
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.767456 | 0.231228 | 0.000431 | 0.000389 | 0.000213 | 0.000288 |