You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000209_00399
You are here: Home > Sequence: MGYG000000209_00399
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
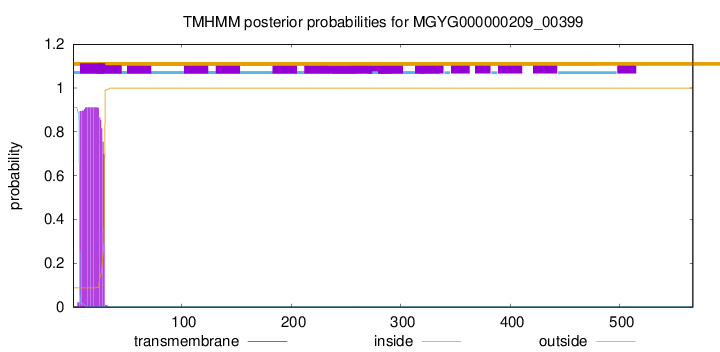
TMHMM annotations
Basic Information help
| Species | Eubacterium_F sp003491505 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Eubacterium_F; Eubacterium_F sp003491505 | |||||||||||
| CAZyme ID | MGYG000000209_00399 | |||||||||||
| CAZy Family | GH10 | |||||||||||
| CAZyme Description | Endo-1,4-beta-xylanase Y | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 86449; End: 88152 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH10 | 171 | 535 | 3.6e-68 | 0.9141914191419142 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00331 | Glyco_hydro_10 | 6.72e-57 | 162 | 532 | 2 | 295 | Glycosyl hydrolase family 10. |
| smart00633 | Glyco_10 | 2.93e-53 | 252 | 532 | 19 | 250 | Glycosyl hydrolase family 10. |
| COG3693 | XynA | 5.52e-39 | 190 | 566 | 47 | 341 | Endo-1,4-beta-xylanase, GH35 family [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| CDM69886.1 | 9.01e-94 | 161 | 565 | 204 | 557 |
| QFJ54853.1 | 5.24e-91 | 157 | 534 | 27 | 367 |
| CBK75021.1 | 5.67e-88 | 168 | 534 | 38 | 367 |
| AFU34339.1 | 9.15e-87 | 168 | 533 | 174 | 504 |
| ADL33480.1 | 9.47e-85 | 173 | 534 | 722 | 1039 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6FHE_A | 1.78e-33 | 195 | 526 | 40 | 312 | Highlyactive enzymes by automated modular backbone assembly and sequence design [synthetic construct] |
| 6D5C_A | 1.07e-32 | 188 | 523 | 42 | 320 | Structureof Caldicellulosiruptor danielii GH10 module of glycoside hydrolase WP_045175321 [Caldicellulosiruptor danielii],6D5C_B Structure of Caldicellulosiruptor danielii GH10 module of glycoside hydrolase WP_045175321 [Caldicellulosiruptor danielii],6D5C_C Structure of Caldicellulosiruptor danielii GH10 module of glycoside hydrolase WP_045175321 [Caldicellulosiruptor danielii] |
| 5OFJ_A | 1.62e-32 | 186 | 523 | 28 | 308 | Crystalstructure of N-terminal domain of bifunctional CbXyn10C [Caldicellulosiruptor bescii DSM 6725] |
| 5OFK_A | 1.06e-31 | 186 | 523 | 28 | 308 | Crystalstructure of CbXyn10C variant E140Q/E248Q complexed with xyloheptaose [Caldicellulosiruptor bescii DSM 6725],5OFL_A Crystal structure of CbXyn10C variant E140Q/E248Q complexed with cellohexaose [Caldicellulosiruptor bescii DSM 6725] |
| 2W5F_A | 1.15e-31 | 188 | 532 | 203 | 511 | ChainA, ENDO-1,4-BETA-XYLANASE Y [Acetivibrio thermocellus],2W5F_B Chain B, ENDO-1,4-BETA-XYLANASE Y [Acetivibrio thermocellus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P23551 | 1.50e-74 | 188 | 533 | 58 | 367 | Endo-1,4-beta-xylanase A OS=Butyrivibrio fibrisolvens OX=831 GN=xynA PE=3 SV=1 |
| P26223 | 4.27e-36 | 182 | 535 | 20 | 324 | Endo-1,4-beta-xylanase B OS=Butyrivibrio fibrisolvens OX=831 GN=xynB PE=3 SV=1 |
| Q60037 | 2.47e-34 | 153 | 534 | 362 | 678 | Endo-1,4-beta-xylanase A OS=Thermotoga maritima (strain ATCC 43589 / DSM 3109 / JCM 10099 / NBRC 100826 / MSB8) OX=243274 GN=xynA PE=1 SV=1 |
| P29126 | 7.27e-34 | 192 | 533 | 651 | 937 | Bifunctional endo-1,4-beta-xylanase XylA OS=Ruminococcus flavefaciens OX=1265 GN=xynA PE=3 SV=1 |
| Q60042 | 4.52e-33 | 154 | 534 | 359 | 674 | Endo-1,4-beta-xylanase A OS=Thermotoga neapolitana OX=2337 GN=xynA PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
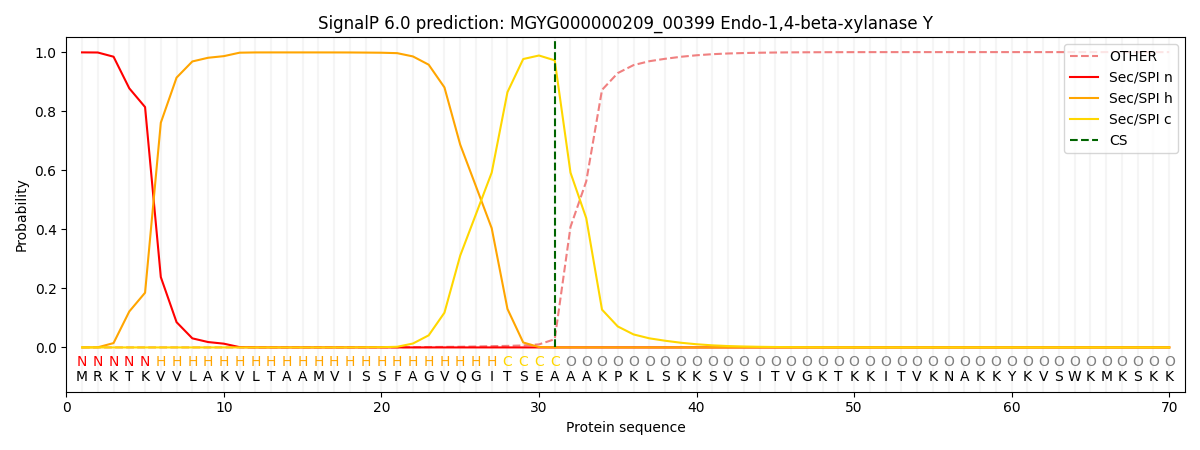
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000415 | 0.997808 | 0.001235 | 0.000200 | 0.000169 | 0.000147 |