You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000209_02018
You are here: Home > Sequence: MGYG000000209_02018
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
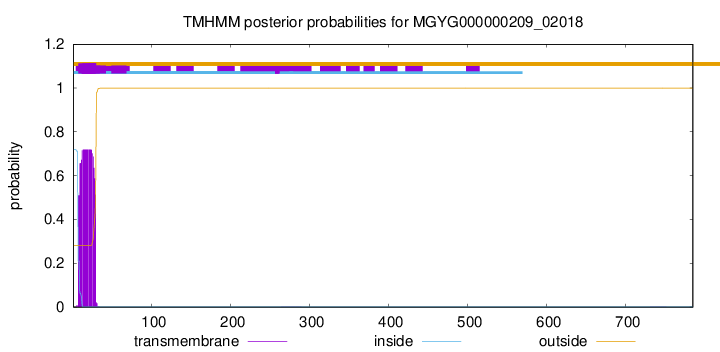
TMHMM annotations
Basic Information help
| Species | Eubacterium_F sp003491505 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Eubacterium_F; Eubacterium_F sp003491505 | |||||||||||
| CAZyme ID | MGYG000000209_02018 | |||||||||||
| CAZy Family | GH10 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 8244; End: 10601 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH10 | 402 | 780 | 3.7e-72 | 0.9702970297029703 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00331 | Glyco_hydro_10 | 3.10e-62 | 397 | 780 | 1 | 310 | Glycosyl hydrolase family 10. |
| smart00633 | Glyco_10 | 3.05e-61 | 439 | 778 | 1 | 263 | Glycosyl hydrolase family 10. |
| COG3693 | XynA | 1.13e-40 | 419 | 783 | 47 | 342 | Endo-1,4-beta-xylanase, GH35 family [Carbohydrate transport and metabolism]. |
| pfam04886 | PT | 2.61e-08 | 146 | 181 | 1 | 36 | PT repeat. This short repeat is composed on the tetrapeptide XPTX. This repeat is found in a variety of proteins, however it is not clear if these repeats are homologous to each other. The alignment represents nine copies of this repeat. |
| pfam04886 | PT | 1.59e-06 | 144 | 177 | 3 | 36 | PT repeat. This short repeat is composed on the tetrapeptide XPTX. This repeat is found in a variety of proteins, however it is not clear if these repeats are homologous to each other. The alignment represents nine copies of this repeat. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ACZ98618.1 | 2.00e-125 | 384 | 782 | 285 | 664 |
| AUZ20632.1 | 6.88e-114 | 394 | 785 | 61 | 436 |
| QUA53142.1 | 7.30e-103 | 385 | 782 | 23 | 405 |
| AHF23772.1 | 1.19e-100 | 394 | 782 | 32 | 405 |
| AHF25185.1 | 1.67e-98 | 394 | 782 | 50 | 423 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7NL2_A | 1.01e-40 | 406 | 780 | 20 | 339 | ChainA, Beta-xylanase [Pseudothermotoga thermarum DSM 5069],7NL2_B Chain B, Beta-xylanase [Pseudothermotoga thermarum DSM 5069] |
| 2W5F_A | 4.27e-39 | 315 | 782 | 97 | 528 | ChainA, ENDO-1,4-BETA-XYLANASE Y [Acetivibrio thermocellus],2W5F_B Chain B, ENDO-1,4-BETA-XYLANASE Y [Acetivibrio thermocellus] |
| 2WYS_A | 4.08e-36 | 315 | 782 | 97 | 528 | ChainA, ENDO-1,4-BETA-XYLANASE Y [Acetivibrio thermocellus],2WYS_B Chain B, ENDO-1,4-BETA-XYLANASE Y [Acetivibrio thermocellus],2WZE_A Chain A, ENDO-1,4-BETA-XYLANASE Y [Acetivibrio thermocellus],2WZE_B Chain B, ENDO-1,4-BETA-XYLANASE Y [Acetivibrio thermocellus] |
| 5OFJ_A | 4.25e-35 | 396 | 744 | 7 | 304 | Crystalstructure of N-terminal domain of bifunctional CbXyn10C [Caldicellulosiruptor bescii DSM 6725] |
| 6D5C_A | 1.34e-34 | 396 | 780 | 19 | 349 | Structureof Caldicellulosiruptor danielii GH10 module of glycoside hydrolase WP_045175321 [Caldicellulosiruptor danielii],6D5C_B Structure of Caldicellulosiruptor danielii GH10 module of glycoside hydrolase WP_045175321 [Caldicellulosiruptor danielii],6D5C_C Structure of Caldicellulosiruptor danielii GH10 module of glycoside hydrolase WP_045175321 [Caldicellulosiruptor danielii] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P51584 | 6.81e-38 | 315 | 782 | 108 | 539 | Endo-1,4-beta-xylanase Y OS=Acetivibrio thermocellus OX=1515 GN=xynY PE=1 SV=1 |
| Q60037 | 1.49e-36 | 352 | 756 | 321 | 665 | Endo-1,4-beta-xylanase A OS=Thermotoga maritima (strain ATCC 43589 / DSM 3109 / JCM 10099 / NBRC 100826 / MSB8) OX=243274 GN=xynA PE=1 SV=1 |
| Q60042 | 8.06e-36 | 352 | 756 | 317 | 661 | Endo-1,4-beta-xylanase A OS=Thermotoga neapolitana OX=2337 GN=xynA PE=1 SV=1 |
| P26223 | 4.48e-34 | 406 | 744 | 11 | 303 | Endo-1,4-beta-xylanase B OS=Butyrivibrio fibrisolvens OX=831 GN=xynB PE=3 SV=1 |
| P23551 | 4.34e-33 | 384 | 745 | 28 | 354 | Endo-1,4-beta-xylanase A OS=Butyrivibrio fibrisolvens OX=831 GN=xynA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
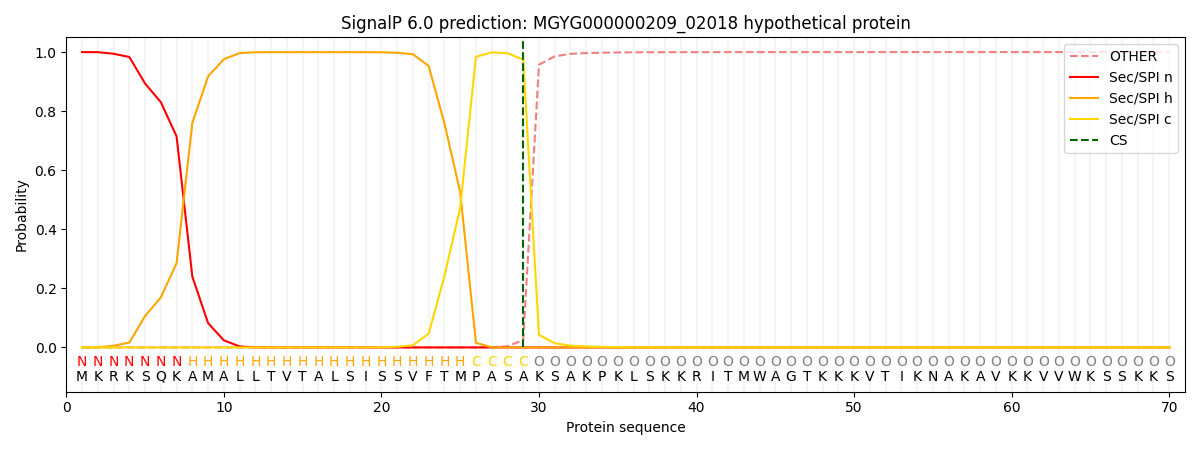
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000266 | 0.999049 | 0.000170 | 0.000194 | 0.000158 | 0.000130 |