You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000209_02587
You are here: Home > Sequence: MGYG000000209_02587
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
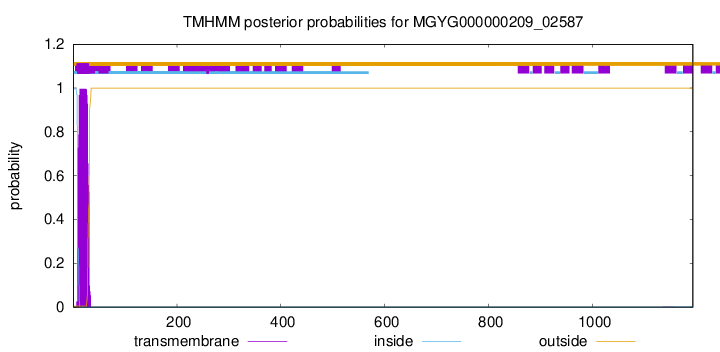
TMHMM annotations
Basic Information help
| Species | Eubacterium_F sp003491505 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Eubacterium_F; Eubacterium_F sp003491505 | |||||||||||
| CAZyme ID | MGYG000000209_02587 | |||||||||||
| CAZy Family | CBM36 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 1116; End: 4697 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH8 | 645 | 956 | 2.7e-83 | 0.9375 |
| CBM36 | 459 | 572 | 1.4e-19 | 0.9565217391304348 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3405 | BcsZ | 4.05e-51 | 600 | 962 | 8 | 349 | Endo-1,4-beta-D-glucanase Y [Carbohydrate transport and metabolism]. |
| PHA03247 | PHA03247 | 1.93e-26 | 155 | 452 | 2667 | 2964 | large tegument protein UL36; Provisional |
| pfam03442 | CBM_X2 | 4.87e-26 | 979 | 1062 | 1 | 83 | Carbohydrate binding domain X2. This domain binds to cellulose and to bacterial cell walls. It is found in glycosyl hydrolases and in scaffolding proteins of cellulosomes (multiprotein glycosyl hydrolase complexes). In the cellulosome it may aid cellulose degradation by anchoring the cellulosome to the bacterial cell wall and by binding it to its substrate. This domain has an Ig-like fold. |
| cd04078 | CBM36_xylanase-like | 1.04e-25 | 457 | 574 | 5 | 118 | Carbohydrate Binding Module family 36 (CBM36); appended mainly to glycoside hydrolase family 11 (GH11) domains; xylan binding. This family includes carbohydrate binding module family 36 (CBM36) most of which appear appended to glycoside hydrolase family 11 (GH11) domains. These CBMs are non-catalytic carbohydrate binding domains that facilitate the strong binding of the GH11 catalytic modules with their dedicated, insoluble substrates. GH11 domains have xylanase (endo-1,4-beta-xylanase) activity which catalyzes the hydrolysis of beta-1,4 bonds of xylan, the major component of hemicelluloses, to generate xylooligosaccharides and xylose. This family includes XynB from Dictyoglomus thermophilum Rt46B.1 and Xyn11A from Pseudobutyrivibrio xylanivorans Mz5T. Xyn11A is a multicatalytic enzyme with an N-terminal GH11 domain, a CBM36 domain, and a C-terminal putative NodB-like polysaccharide deacetylase which is predicted to be an acetyl esterase involved in debranching activity in the xylan backbone. CBM6 is an unusual CBM as it represents a chimera of two distinct binding sites with different modes of binding: binding site I within the loop regions and binding site II on the concave face of the beta-sandwich fold. Consistent with its structural and sequence similarity to CBM6, CBM36 binds xylan, but only at binding site I, and in a calcium-dependent manner; the latter suggests its potential application in affinity labeling. |
| PHA03247 | PHA03247 | 5.59e-25 | 154 | 451 | 2619 | 2954 | large tegument protein UL36; Provisional |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QUL54973.1 | 6.15e-188 | 457 | 1003 | 36 | 578 |
| AAS85781.1 | 2.34e-159 | 588 | 1179 | 650 | 1294 |
| QNU67295.1 | 1.08e-153 | 584 | 1013 | 22 | 448 |
| ALQ07930.1 | 2.06e-149 | 476 | 968 | 47 | 554 |
| QWG04463.1 | 4.96e-146 | 454 | 973 | 45 | 572 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6G00_A | 2.88e-114 | 591 | 965 | 3 | 395 | CrystalStructure of a GH8 xylanase from Teredinibacter turnerae [Teredinibacter turnerae T7901],6G09_A Crystal Structure of a GH8 xylobiose complex from Teredinibacter turnerae [Teredinibacter turnerae T7901],6G0B_A Crystal Structure of a GH8 xylotriose complex from Teredinibacter Turnerae [Teredinibacter turnerae T7901] |
| 6G0N_A | 1.52e-113 | 591 | 965 | 3 | 395 | CrystalStructure of a GH8 catalytic mutant xylohexaose complex xylanase from Teredinibacter turnerae [Teredinibacter turnerae T7901] |
| 6SUD_A | 6.34e-113 | 593 | 964 | 7 | 380 | Structureof L320A mutant of Rex8A from Paenibacillus barcinonensis complexed with xylose. [Paenibacillus barcinonensis],6SUD_B Structure of L320A mutant of Rex8A from Paenibacillus barcinonensis complexed with xylose. [Paenibacillus barcinonensis] |
| 6SHY_A | 1.23e-112 | 593 | 964 | 7 | 380 | Structureof L320A/H321S double mutant of Rex8A from Paenibacillus barcinonensis [Paenibacillus barcinonensis],6SHY_B Structure of L320A/H321S double mutant of Rex8A from Paenibacillus barcinonensis [Paenibacillus barcinonensis] |
| 6SRD_A | 1.23e-112 | 593 | 964 | 7 | 380 | Structureof Rex8A from Paenibacillus barcinonensis complexed with xylose. [Paenibacillus barcinonensis],6SRD_B Structure of Rex8A from Paenibacillus barcinonensis complexed with xylose. [Paenibacillus barcinonensis] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| A0A0S2UQQ5 | 6.31e-112 | 593 | 964 | 7 | 380 | Reducing-end xylose-releasing exo-oligoxylanase Rex8A OS=Paenibacillus barcinonensis OX=198119 GN=rex8A PE=1 SV=1 |
| Q9KB30 | 2.39e-100 | 593 | 964 | 7 | 379 | Reducing end xylose-releasing exo-oligoxylanase OS=Alkalihalobacillus halodurans (strain ATCC BAA-125 / DSM 18197 / FERM 7344 / JCM 9153 / C-125) OX=272558 GN=BH2105 PE=1 SV=1 |
| A1A048 | 2.00e-74 | 602 | 962 | 14 | 377 | Reducing end xylose-releasing exo-oligoxylanase OS=Bifidobacterium adolescentis (strain ATCC 15703 / DSM 20083 / NCTC 11814 / E194a) OX=367928 GN=xylA PE=1 SV=1 |
| P37701 | 1.76e-29 | 652 | 997 | 95 | 427 | Endoglucanase 2 OS=Ruminiclostridium josui OX=1499 GN=celB PE=3 SV=1 |
| Q3MUH7 | 3.43e-15 | 967 | 1093 | 773 | 896 | Xyloglucanase OS=Paenibacillus sp. OX=58172 GN=xeg74 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
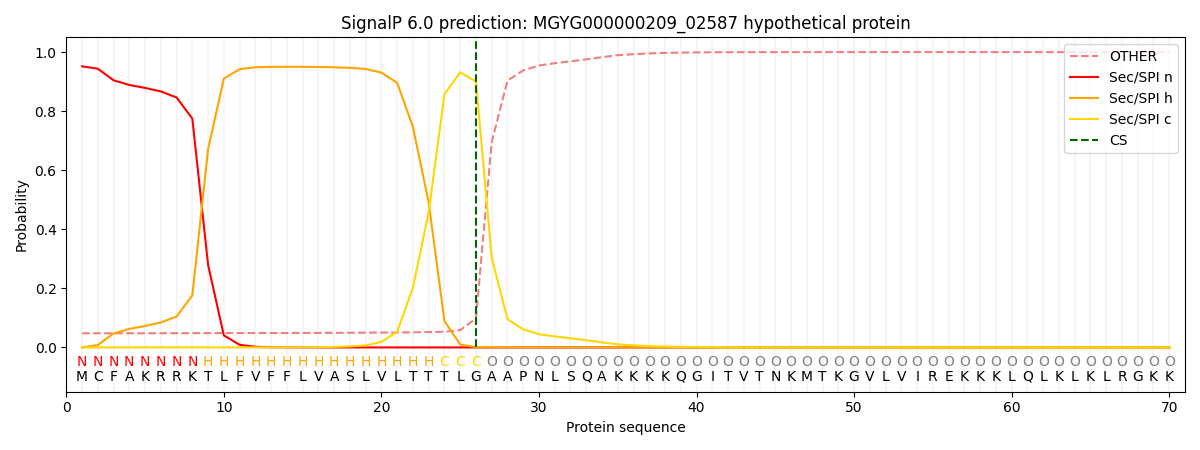
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.053120 | 0.945316 | 0.000587 | 0.000417 | 0.000276 | 0.000261 |