You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000211_00360
You are here: Home > Sequence: MGYG000000211_00360
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
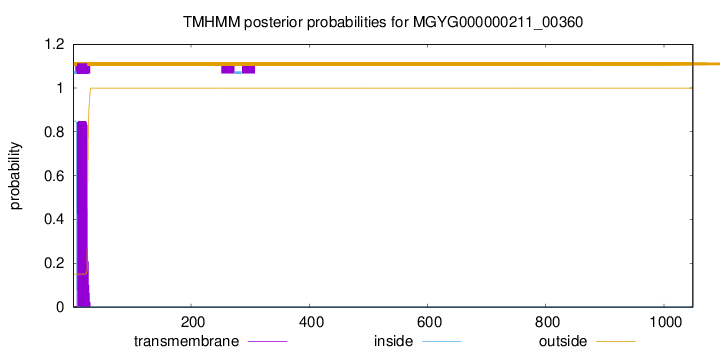
TMHMM annotations
Basic Information help
| Species | Bacteroides sp900556215 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; Bacteroides sp900556215 | |||||||||||
| CAZyme ID | MGYG000000211_00360 | |||||||||||
| CAZy Family | GH3 | |||||||||||
| CAZyme Description | Xylan 1,4-beta-xylosidase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 549134; End: 552283 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH3 | 62 | 303 | 7.2e-70 | 0.9768518518518519 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| PRK15098 | PRK15098 | 6.01e-134 | 4 | 844 | 1 | 758 | beta-glucosidase BglX. |
| PLN03080 | PLN03080 | 2.05e-121 | 34 | 837 | 48 | 769 | Probable beta-xylosidase; Provisional |
| COG1472 | BglX | 2.90e-75 | 43 | 435 | 35 | 392 | Periplasmic beta-glucosidase and related glycosidases [Carbohydrate transport and metabolism]. |
| cd01827 | sialate_O-acetylesterase_like1 | 4.93e-60 | 855 | 1040 | 1 | 184 | sialate O-acetylesterase_like family of the SGNH hydrolases, a diverse family of lipases and esterases. The tertiary fold of the enzyme is substantially different from that of the alpha/beta hydrolase family and unique among all known hydrolases; its active site closely resembles the Ser-His-Asp(Glu) triad found in other serine hydrolases. |
| pfam00933 | Glyco_hydro_3 | 6.67e-54 | 42 | 336 | 42 | 316 | Glycosyl hydrolase family 3 N terminal domain. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ALJ59400.1 | 0.0 | 1 | 854 | 1 | 854 |
| QUT89559.1 | 0.0 | 1 | 854 | 1 | 854 |
| QDO67855.1 | 0.0 | 1 | 854 | 1 | 854 |
| QRQ47317.1 | 0.0 | 1 | 854 | 1 | 855 |
| QUT46491.1 | 0.0 | 1 | 854 | 1 | 855 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5Z87_A | 1.58e-90 | 63 | 844 | 119 | 778 | ChainA, EmGH1 [Aurantiacibacter marinus],5Z87_B Chain B, EmGH1 [Aurantiacibacter marinus] |
| 3AC0_A | 6.94e-89 | 39 | 843 | 6 | 828 | Crystalstructure of Beta-glucosidase from Kluyveromyces marxianus in complex with glucose [Kluyveromyces marxianus],3AC0_B Crystal structure of Beta-glucosidase from Kluyveromyces marxianus in complex with glucose [Kluyveromyces marxianus],3AC0_C Crystal structure of Beta-glucosidase from Kluyveromyces marxianus in complex with glucose [Kluyveromyces marxianus],3AC0_D Crystal structure of Beta-glucosidase from Kluyveromyces marxianus in complex with glucose [Kluyveromyces marxianus] |
| 6R5I_A | 5.61e-86 | 63 | 842 | 68 | 724 | ChainA, Periplasmic beta-glucosidase [Pseudomonas aeruginosa PAO1],6R5I_B Chain B, Periplasmic beta-glucosidase [Pseudomonas aeruginosa PAO1],6R5N_A Chain A, Periplasmic beta-glucosidase [Pseudomonas aeruginosa PAO1],6R5N_B Chain B, Periplasmic beta-glucosidase [Pseudomonas aeruginosa PAO1] |
| 6R5O_A | 2.79e-85 | 63 | 842 | 68 | 724 | ChainA, Periplasmic beta-glucosidase [Pseudomonas aeruginosa PAO1],6R5O_B Chain B, Periplasmic beta-glucosidase [Pseudomonas aeruginosa PAO1],6R5R_B Chain B, Periplasmic beta-glucosidase [Pseudomonas aeruginosa PAO1],6R5T_A Chain A, Periplasmic beta-glucosidase [Pseudomonas aeruginosa PAO1],6R5T_B Chain B, Periplasmic beta-glucosidase [Pseudomonas aeruginosa PAO1],6R5U_A Chain A, Periplasmic beta-glucosidase [Pseudomonas aeruginosa PAO1],6R5U_B Chain B, Periplasmic beta-glucosidase [Pseudomonas aeruginosa PAO1],6R5V_A Chain A, Periplasmic beta-glucosidase [Pseudomonas aeruginosa PAO1],6R5V_B Chain B, Periplasmic beta-glucosidase [Pseudomonas aeruginosa PAO1] |
| 6R5R_A | 2.85e-85 | 63 | 842 | 69 | 725 | ChainA, Periplasmic beta-glucosidase [Pseudomonas aeruginosa PAO1] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| D5EY15 | 7.79e-152 | 13 | 847 | 7 | 856 | Xylan 1,4-beta-xylosidase OS=Prevotella ruminicola (strain ATCC 19189 / JCM 8958 / 23) OX=264731 GN=xyl3A PE=1 SV=1 |
| T2KMH0 | 5.00e-108 | 4 | 848 | 1 | 718 | Beta-xylosidase OS=Formosa agariphila (strain DSM 15362 / KCTC 12365 / LMG 23005 / KMM 3901 / M-2Alg 35-1) OX=1347342 GN=BN863_22130 PE=1 SV=1 |
| Q94KD8 | 8.05e-106 | 34 | 846 | 50 | 759 | Probable beta-D-xylosidase 2 OS=Arabidopsis thaliana OX=3702 GN=BXL2 PE=2 SV=1 |
| Q9SGZ5 | 2.10e-102 | 34 | 843 | 44 | 763 | Probable beta-D-xylosidase 7 OS=Arabidopsis thaliana OX=3702 GN=BXL7 PE=2 SV=2 |
| Q9LXD6 | 8.37e-97 | 38 | 845 | 59 | 771 | Beta-D-xylosidase 3 OS=Arabidopsis thaliana OX=3702 GN=BXL3 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
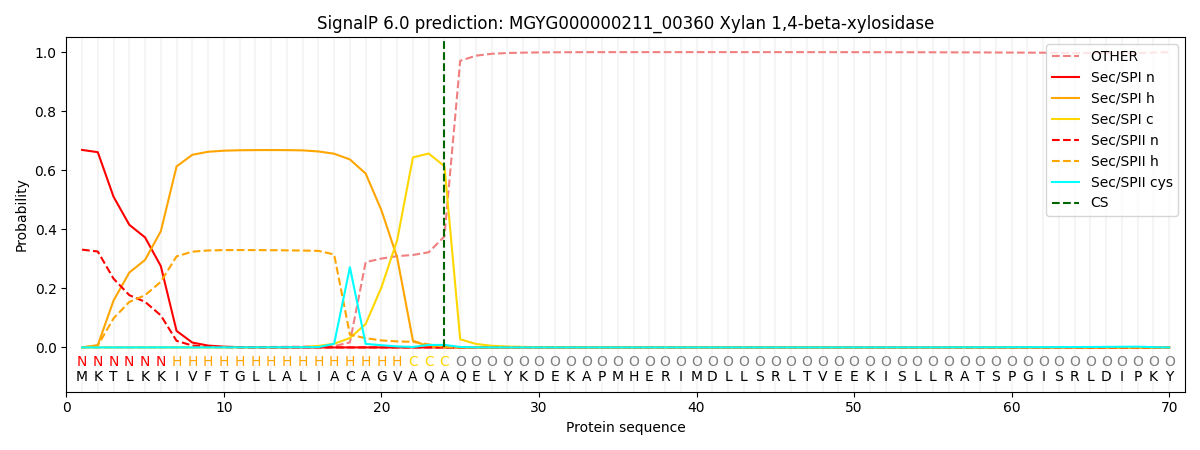
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000954 | 0.655060 | 0.342979 | 0.000337 | 0.000337 | 0.000308 |