You are browsing environment: HUMAN GUT
MGYG000000211_00831
Basic Information
help
Species
Bacteroides sp900556215
Lineage
Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; Bacteroides sp900556215
CAZyme ID
MGYG000000211_00831
CAZy Family
GH5
CAZyme Description
hypothetical protein
CAZyme Property
Genome Property
Genome Assembly ID
Genome Size
Genome Type
Country
Continent
MGYG000000211
6486320
Isolate
China
Asia
Gene Location
Start: 355082;
End: 357046
Strand: -
No EC number prediction in MGYG000000211_00831.
Family
Start
End
Evalue
family coverage
GH5
69
349
9.7e-111
0.9884169884169884
Cdd ID
Domain
E-Value
qStart
qEnd
sStart
sEnd
Domain Description
pfam00150
Cellulase
5.13e-08
63
306
2
208
Cellulase (glycosyl hydrolase family 5).
more
PRK10819
PRK10819
0.001
25
53
60
92
transport protein TonB; Provisional
more
pfam16070
TMEM132
0.005
457
523
17
84
Transmembrane protein family 132. This presumed domain is found in members of the TMEM132 family. TMEM132A may be involved in embryonic and postnatal brain development. TMEM132D may be a marker for oligodendrocyte differentiation.
more
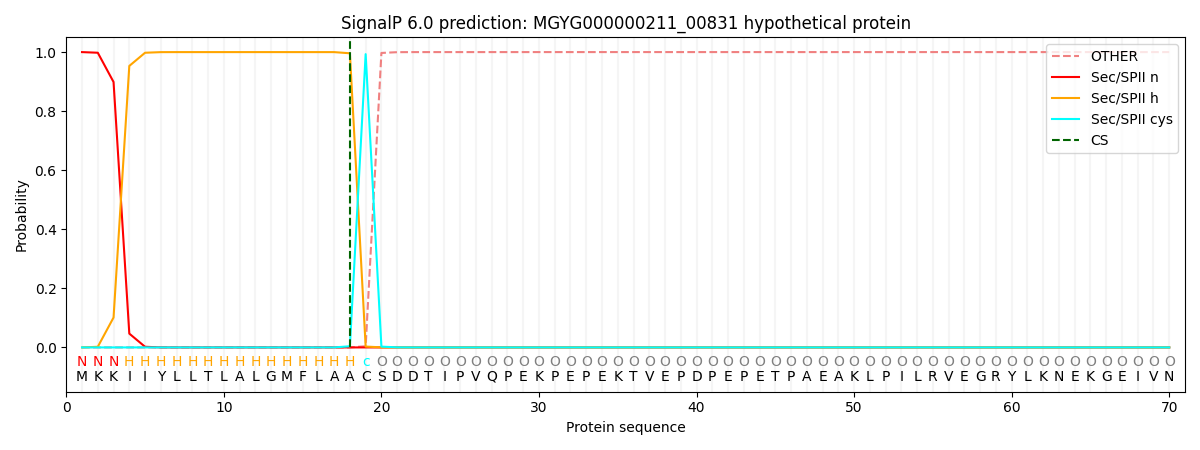
This protein is predicted as LIPO
Other
SP_Sec_SPI
LIPO_Sec_SPII
TAT_Tat_SPI
TATLIP_Sec_SPII
PILIN_Sec_SPIII
0.000000
0.000000
1.000039
0.000000
0.000000
0.000000
There is no transmembrane helices in MGYG000000211_00831.