You are browsing environment: HUMAN GUT
MGYG000000211_02897
Basic Information
help
Species
Bacteroides sp900556215
Lineage
Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; Bacteroides sp900556215
CAZyme ID
MGYG000000211_02897
CAZy Family
GH28
CAZyme Description
hypothetical protein
CAZyme Property
Protein Length
CGC
Molecular Weight
Isoelectric Point
476
53156.48
7.4335
Genome Property
Genome Assembly ID
Genome Size
Genome Type
Country
Continent
MGYG000000211
6486320
Isolate
China
Asia
Gene Location
Start: 129809;
End: 131239
Strand: -
No EC number prediction in MGYG000000211_02897.
Family
Start
End
Evalue
family coverage
GH28
177
442
2.5e-32
0.7784615384615384
Cdd ID
Domain
E-Value
qStart
qEnd
sStart
sEnd
Domain Description
pfam00295
Glyco_hydro_28
0.005
219
414
88
279
Glycosyl hydrolases family 28. Glycosyl hydrolase family 28 includes polygalacturonase EC:3.2.1.15 as well as rhamnogalacturonase A(RGase A), EC:3.2.1.-. These enzymes are important in cell wall metabolism.
more
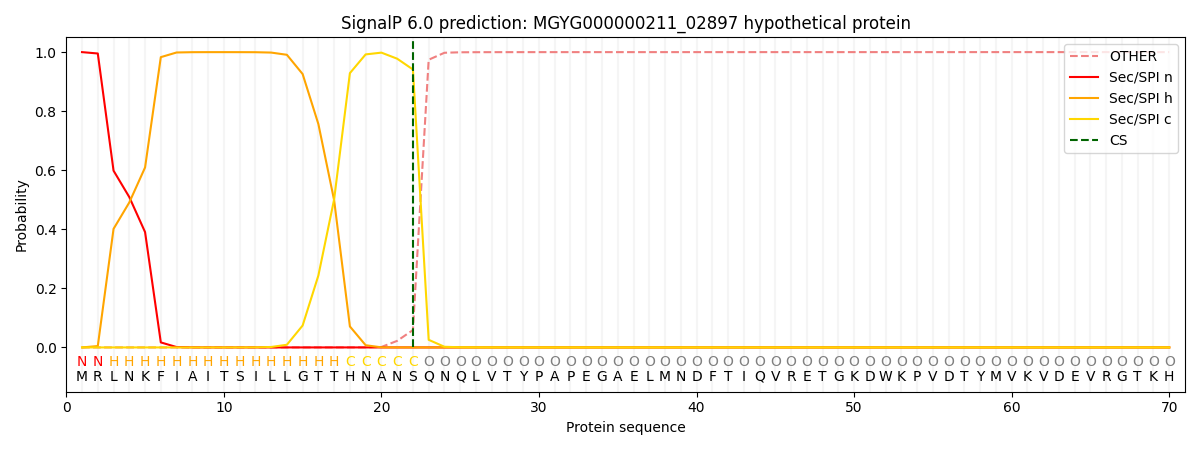
This protein is predicted as SP
Other
SP_Sec_SPI
LIPO_Sec_SPII
TAT_Tat_SPI
TATLIP_Sec_SPII
PILIN_Sec_SPIII
0.000503
0.998589
0.000291
0.000206
0.000179
0.000171
There is no transmembrane helices in MGYG000000211_02897.