You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000211_04562
You are here: Home > Sequence: MGYG000000211_04562
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
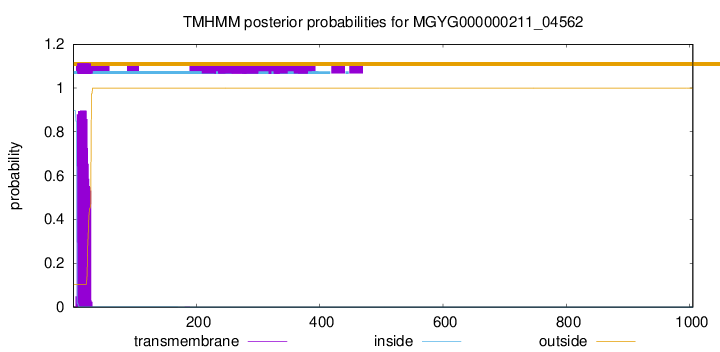
TMHMM annotations
Basic Information help
| Species | Bacteroides sp900556215 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; Bacteroides sp900556215 | |||||||||||
| CAZyme ID | MGYG000000211_04562 | |||||||||||
| CAZy Family | GH55 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 72388; End: 75405 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH55 | 42 | 674 | 4.6e-86 | 0.8554054054054054 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam08450 | SGL | 5.29e-13 | 906 | 994 | 145 | 243 | SMP-30/Gluconolaconase/LRE-like region. This family describes a region that is found in proteins expressed by a variety of eukaryotic and prokaryotic species. These proteins include various enzymes, such as senescence marker protein 30 (SMP-30), gluconolactonase and luciferin-regenerating enzyme (LRE). SMP-30 is known to hydrolyze diisopropyl phosphorofluoridate in the liver, and has been noted as having sequence similarity, in the region described in this family, with PON1 and LRE. |
| COG3386 | YvrE | 6.59e-13 | 719 | 995 | 17 | 274 | Sugar lactone lactonase YvrE [Carbohydrate transport and metabolism]. |
| pfam12708 | Pectate_lyase_3 | 3.11e-10 | 46 | 218 | 7 | 171 | Pectate lyase superfamily protein. This family of proteins possesses a beta helical structure like Pectate lyase. This family is most closely related to glycosyl hydrolase family 28. |
| pfam12708 | Pectate_lyase_3 | 1.66e-09 | 379 | 432 | 1 | 60 | Pectate lyase superfamily protein. This family of proteins possesses a beta helical structure like Pectate lyase. This family is most closely related to glycosyl hydrolase family 28. |
| COG5434 | Pgu1 | 1.21e-07 | 319 | 543 | 25 | 244 | Polygalacturonase [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QUU00135.1 | 0.0 | 1 | 1005 | 1 | 1005 |
| QMI79027.1 | 0.0 | 1 | 1005 | 1 | 1004 |
| QBJ17509.1 | 0.0 | 1 | 1005 | 1 | 1004 |
| QPH57852.1 | 0.0 | 1 | 1005 | 1 | 1004 |
| BBK89258.1 | 0.0 | 1 | 1005 | 1 | 1004 |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
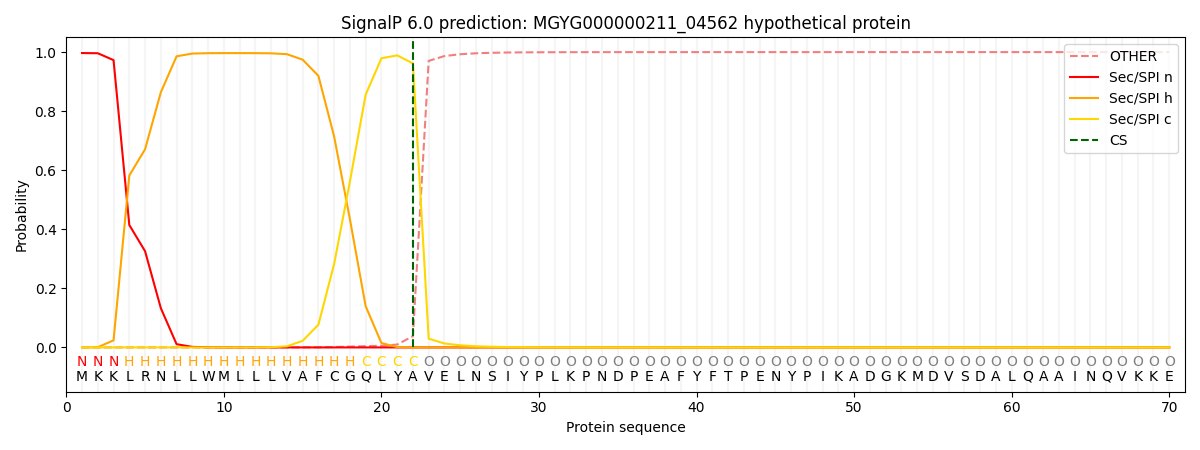
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000617 | 0.994793 | 0.003957 | 0.000211 | 0.000202 | 0.000196 |