You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000214_01055
You are here: Home > Sequence: MGYG000000214_01055
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Veillonella dispar_A | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_C; Negativicutes; Veillonellales; Veillonellaceae; Veillonella; Veillonella dispar_A | |||||||||||
| CAZyme ID | MGYG000000214_01055 | |||||||||||
| CAZy Family | GT83 | |||||||||||
| CAZyme Description | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 26194; End: 27822 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT83 | 7 | 486 | 9.2e-96 | 0.9259259259259259 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG1807 | ArnT | 2.58e-51 | 9 | 368 | 12 | 376 | 4-amino-4-deoxy-L-arabinose transferase or related glycosyltransferase of PMT family [Cell wall/membrane/envelope biogenesis]. |
| PRK13279 | arnT | 9.35e-29 | 1 | 366 | 2 | 371 | lipid IV(A) 4-amino-4-deoxy-L-arabinosyltransferase. |
| pfam13231 | PMT_2 | 4.29e-20 | 60 | 219 | 1 | 157 | Dolichyl-phosphate-mannose-protein mannosyltransferase. This family contains members that are not captured by pfam02366. |
| COG4745 | COG4745 | 2.86e-04 | 55 | 339 | 56 | 383 | Predicted membrane-bound mannosyltransferase [General function prediction only]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BBU37212.1 | 0.0 | 1 | 542 | 1 | 542 |
| QQB17174.1 | 0.0 | 1 | 542 | 1 | 542 |
| SNV02078.1 | 0.0 | 1 | 542 | 1 | 542 |
| ACZ25367.1 | 0.0 | 1 | 542 | 1 | 542 |
| CAB1277318.1 | 0.0 | 1 | 542 | 1 | 542 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5EZM_A | 7.98e-30 | 9 | 312 | 40 | 348 | CrystalStructure of ArnT from Cupriavidus metallidurans in the apo state [Cupriavidus metallidurans CH34],5F15_A Crystal Structure of ArnT from Cupriavidus metallidurans bound to Undecaprenyl phosphate [Cupriavidus metallidurans CH34] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| B4TBG8 | 2.45e-26 | 17 | 335 | 18 | 337 | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase OS=Salmonella heidelberg (strain SL476) OX=454169 GN=arnT PE=3 SV=1 |
| Q57M54 | 4.80e-24 | 17 | 335 | 17 | 336 | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase OS=Salmonella choleraesuis (strain SC-B67) OX=321314 GN=arnT PE=3 SV=2 |
| Q8Z538 | 2.06e-23 | 17 | 335 | 17 | 336 | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase OS=Salmonella typhi OX=90370 GN=arnT PE=3 SV=2 |
| O52327 | 2.06e-23 | 17 | 335 | 17 | 336 | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase OS=Salmonella typhimurium (strain LT2 / SGSC1412 / ATCC 700720) OX=99287 GN=arnT PE=1 SV=2 |
| Q5PNA8 | 2.06e-23 | 17 | 335 | 17 | 336 | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase OS=Salmonella paratyphi A (strain ATCC 9150 / SARB42) OX=295319 GN=arnT PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.999615 | 0.000393 | 0.000005 | 0.000001 | 0.000001 | 0.000002 |
TMHMM Annotations download full data without filtering help
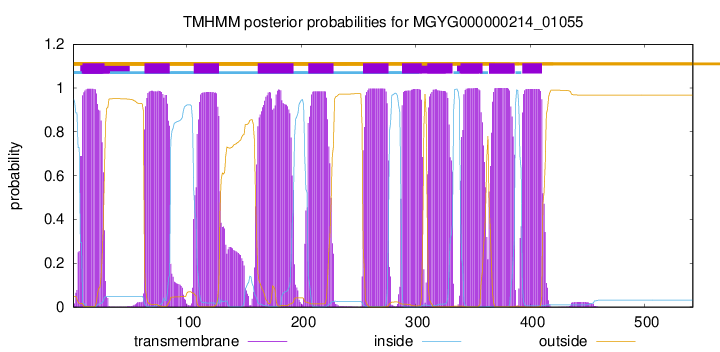
| start | end |
|---|---|
| 9 | 28 |
| 63 | 85 |
| 106 | 128 |
| 162 | 193 |
| 206 | 228 |
| 254 | 276 |
| 288 | 305 |
| 310 | 332 |
| 339 | 358 |
| 364 | 386 |
| 393 | 410 |
