You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000215_01095
You are here: Home > Sequence: MGYG000000215_01095
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Prevotella stercorea | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Prevotella; Prevotella stercorea | |||||||||||
| CAZyme ID | MGYG000000215_01095 | |||||||||||
| CAZy Family | GH92 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 26298; End: 27080 Strand: + | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam17678 | Glyco_hydro_92N | 1.23e-64 | 28 | 244 | 1 | 211 | Glycosyl hydrolase family 92 N-terminal domain. This domain is found at the N-terminus of family 92 glycosyl hydrolase proteins. |
| COG3537 | COG3537 | 2.23e-20 | 8 | 228 | 19 | 239 | Putative alpha-1,2-mannosidase [Carbohydrate transport and metabolism]. |
| NF035929 | lectin_1 | 1.74e-19 | 29 | 201 | 11 | 171 | lectin. Lectins are important adhesin proteins, which bind carbohydrate structures on host cell surface. The carbohydrate specificity of diverse lectins to a large extent dictates bacteria tissue tropism by mediating specific attachment to unique host sites expressing the corresponding carbohydrate receptor. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ADE83124.1 | 9.57e-128 | 16 | 249 | 15 | 248 |
| QVJ80984.1 | 8.28e-126 | 16 | 249 | 15 | 248 |
| QYR10970.1 | 1.65e-122 | 22 | 249 | 21 | 248 |
| ALO49805.1 | 2.70e-121 | 22 | 249 | 6 | 233 |
| AGB28393.1 | 8.52e-116 | 5 | 249 | 3 | 249 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6F91_A | 4.09e-21 | 20 | 219 | 2 | 199 | Structureof the family GH92 alpha-mannosidase BT3965 from Bacteroides thetaiotaomicron [Bacteroides thetaiotaomicron],6F91_B Structure of the family GH92 alpha-mannosidase BT3965 from Bacteroides thetaiotaomicron [Bacteroides thetaiotaomicron],6F91_C Structure of the family GH92 alpha-mannosidase BT3965 from Bacteroides thetaiotaomicron [Bacteroides thetaiotaomicron],6F91_D Structure of the family GH92 alpha-mannosidase BT3965 from Bacteroides thetaiotaomicron [Bacteroides thetaiotaomicron],6F91_E Structure of the family GH92 alpha-mannosidase BT3965 from Bacteroides thetaiotaomicron [Bacteroides thetaiotaomicron],6F91_F Structure of the family GH92 alpha-mannosidase BT3965 from Bacteroides thetaiotaomicron [Bacteroides thetaiotaomicron],6F91_G Structure of the family GH92 alpha-mannosidase BT3965 from Bacteroides thetaiotaomicron [Bacteroides thetaiotaomicron],6F91_H Structure of the family GH92 alpha-mannosidase BT3965 from Bacteroides thetaiotaomicron [Bacteroides thetaiotaomicron],6F92_A Structure of the family GH92 alpha-mannosidase BT3965 from Bacteroides thetaiotaomicron in complex with Mannoimidazole (ManI) [Bacteroides thetaiotaomicron],6F92_B Structure of the family GH92 alpha-mannosidase BT3965 from Bacteroides thetaiotaomicron in complex with Mannoimidazole (ManI) [Bacteroides thetaiotaomicron],6F92_C Structure of the family GH92 alpha-mannosidase BT3965 from Bacteroides thetaiotaomicron in complex with Mannoimidazole (ManI) [Bacteroides thetaiotaomicron],6F92_D Structure of the family GH92 alpha-mannosidase BT3965 from Bacteroides thetaiotaomicron in complex with Mannoimidazole (ManI) [Bacteroides thetaiotaomicron] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
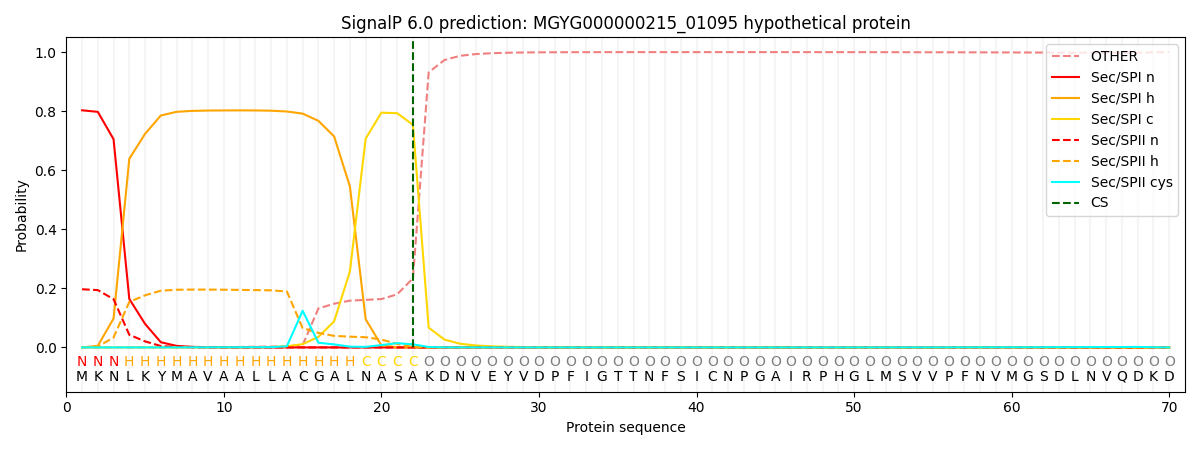
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000874 | 0.791394 | 0.206806 | 0.000347 | 0.000289 | 0.000262 |
