You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000218_00349
You are here: Home > Sequence: MGYG000000218_00349
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Acidaminococcus provencensis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_C; Negativicutes; Acidaminococcales; Acidaminococcaceae; Acidaminococcus; Acidaminococcus provencensis | |||||||||||
| CAZyme ID | MGYG000000218_00349 | |||||||||||
| CAZy Family | GH73 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 125111; End: 126298 Strand: - | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam01832 | Glucosaminidase | 1.21e-09 | 251 | 323 | 1 | 84 | Mannosyl-glycoprotein endo-beta-N-acetylglucosaminidase. This family includes Mannosyl-glycoprotein endo-beta-N-acetylglucosaminidase EC:3.2.1.96. As well as the flageller protein J that has been shown to hydrolyze peptidoglycan. |
| PTZ00121 | PTZ00121 | 1.56e-04 | 28 | 191 | 1646 | 1802 | MAEBL; Provisional |
| PTZ00121 | PTZ00121 | 1.60e-04 | 29 | 138 | 1269 | 1389 | MAEBL; Provisional |
| NF012221 | MARTX_Nterm | 0.005 | 44 | 170 | 1710 | 1838 | MARTX multifunctional-autoprocessing repeats-in-toxin holotoxin N-terminal region. This model describes the N-terminal 1900 amino acids of MARTX family multifunctional-autoprocessing repeats-in-toxin holotoxins, which contain both repeat regions that facilitate their entry into eukaryotic target cells, and multiple effector domains. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ADB47590.1 | 3.07e-108 | 160 | 390 | 151 | 381 |
| AEQ22681.1 | 1.70e-97 | 180 | 389 | 84 | 293 |
| QNP78284.1 | 4.13e-61 | 210 | 385 | 126 | 301 |
| BBG63749.1 | 7.14e-61 | 210 | 385 | 146 | 321 |
| SNV01267.1 | 1.56e-52 | 211 | 375 | 98 | 262 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| O51481 | 5.89e-20 | 250 | 375 | 71 | 191 | Uncharacterized protein BB_0531 OS=Borreliella burgdorferi (strain ATCC 35210 / DSM 4680 / CIP 102532 / B31) OX=224326 GN=BB_0531 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
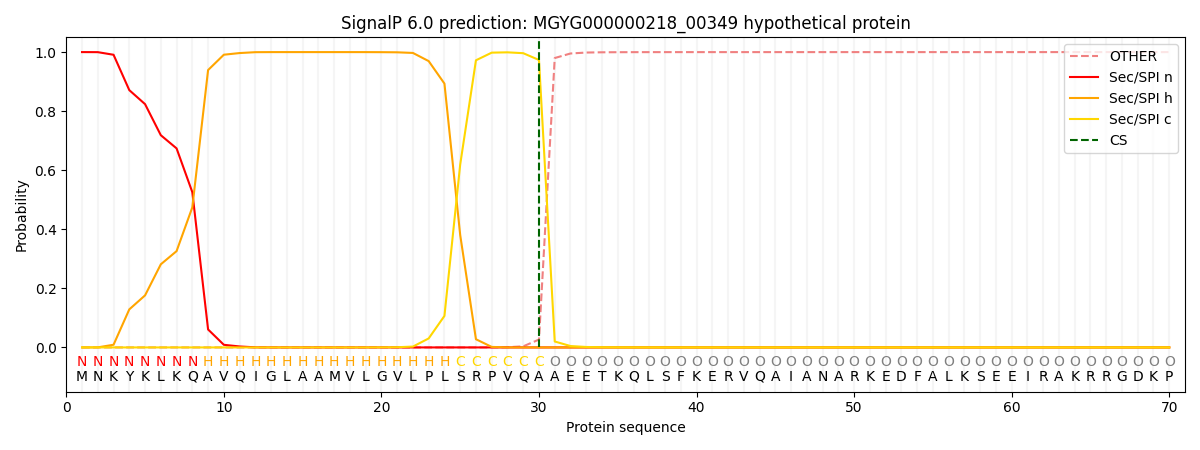
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000286 | 0.999045 | 0.000198 | 0.000165 | 0.000155 | 0.000143 |
