You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000219_03267
You are here: Home > Sequence: MGYG000000219_03267
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
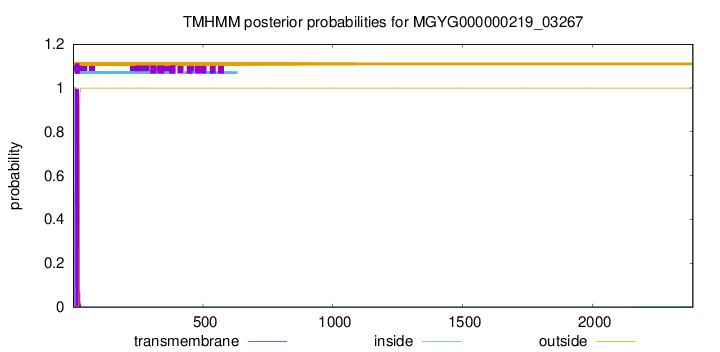
TMHMM annotations
Basic Information help
| Species | Eubacterium maltosivorans | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Eubacteriales; Eubacteriaceae; Eubacterium; Eubacterium maltosivorans | |||||||||||
| CAZyme ID | MGYG000000219_03267 | |||||||||||
| CAZy Family | CBM54 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 52513; End: 59676 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM54 | 32 | 137 | 3e-22 | 0.9385964912280702 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG5492 | YjdB | 5.78e-22 | 386 | 548 | 181 | 328 | Uncharacterized conserved protein YjdB, contains Ig-like domain [General function prediction only]. |
| COG5492 | YjdB | 1.89e-16 | 863 | 956 | 167 | 261 | Uncharacterized conserved protein YjdB, contains Ig-like domain [General function prediction only]. |
| pfam02368 | Big_2 | 2.18e-12 | 878 | 954 | 1 | 77 | Bacterial Ig-like domain (group 2). This family consists of bacterial domains with an Ig-like fold. Members of this family are found in bacterial and phage surface proteins such as intimins. |
| pfam02368 | Big_2 | 6.19e-12 | 387 | 463 | 1 | 77 | Bacterial Ig-like domain (group 2). This family consists of bacterial domains with an Ig-like fold. Members of this family are found in bacterial and phage surface proteins such as intimins. |
| pfam02368 | Big_2 | 4.70e-09 | 476 | 547 | 1 | 72 | Bacterial Ig-like domain (group 2). This family consists of bacterial domains with an Ig-like fold. Members of this family are found in bacterial and phage surface proteins such as intimins. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QCT72360.1 | 0.0 | 1 | 2387 | 1 | 2373 |
| QSW19407.1 | 1.99e-27 | 384 | 587 | 1797 | 2008 |
| ART28007.1 | 4.74e-26 | 386 | 973 | 1296 | 1884 |
| ADL50747.1 | 1.75e-22 | 384 | 556 | 199 | 369 |
| CBL09766.1 | 5.76e-22 | 384 | 631 | 1444 | 1667 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P35838 | 1.12e-08 | 878 | 956 | 237 | 316 | Uncharacterized protein CA_C0552 OS=Clostridium acetobutylicum (strain ATCC 824 / DSM 792 / JCM 1419 / LMG 5710 / VKM B-1787) OX=272562 GN=CA_C0552 PE=4 SV=2 |
| P85991 | 5.04e-08 | 837 | 956 | 139 | 257 | Ig-like virion protein OS=Serratia phage KSP90 OX=552528 PE=1 SV=2 |
| P33747 | 5.32e-08 | 878 | 967 | 38 | 128 | Uncharacterized protein CA_P0160 OS=Clostridium acetobutylicum (strain ATCC 824 / DSM 792 / JCM 1419 / LMG 5710 / VKM B-1787) OX=272562 GN=CA_P0160 PE=3 SV=2 |
| A0A3R0A696 | 2.78e-07 | 870 | 1118 | 275 | 549 | Alpha-L-arabinofuranosidase OS=Bifidobacterium longum subsp. longum (strain ATCC 15707 / DSM 20219 / JCM 1217 / NCTC 11818 / E194b) OX=565042 GN=blArafA PE=1 SV=1 |
| Q18DN4 | 1.34e-06 | 2005 | 2147 | 4336 | 4485 | Halomucin OS=Haloquadratum walsbyi (strain DSM 16790 / HBSQ001) OX=362976 GN=hmu PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
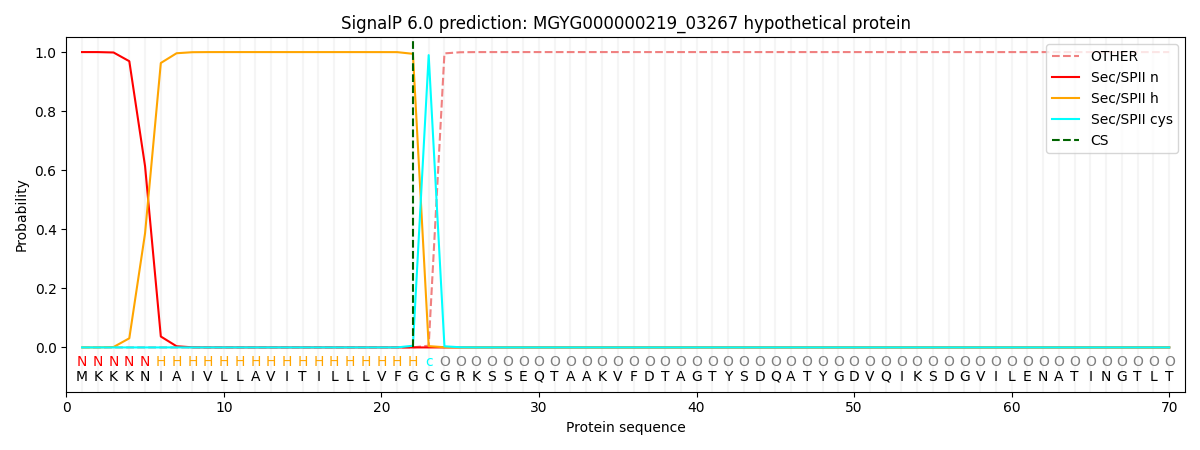
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000001 | 1.000069 | 0.000000 | 0.000000 | 0.000000 |