You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000221_01970
You are here: Home > Sequence: MGYG000000221_01970
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Parabacteroides bouchesdurhonensis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Tannerellaceae; Parabacteroides; Parabacteroides bouchesdurhonensis | |||||||||||
| CAZyme ID | MGYG000000221_01970 | |||||||||||
| CAZy Family | CE15 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 180700; End: 183033 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE15 | 85 | 403 | 2.3e-56 | 0.9814126394052045 |
| CE19 | 515 | 751 | 8.1e-18 | 0.6144578313253012 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG1506 | DAP2 | 2.48e-12 | 520 | 671 | 380 | 516 | Dipeptidyl aminopeptidase/acylaminoacyl peptidase [Amino acid transport and metabolism]. |
| COG0657 | Aes | 1.13e-06 | 466 | 717 | 13 | 253 | Acetyl esterase/lipase [Lipid transport and metabolism]. |
| pfam00561 | Abhydrolase_1 | 2.37e-06 | 536 | 664 | 1 | 106 | alpha/beta hydrolase fold. This catalytic domain is found in a very wide range of enzymes. |
| COG1506 | DAP2 | 9.33e-06 | 166 | 282 | 382 | 507 | Dipeptidyl aminopeptidase/acylaminoacyl peptidase [Amino acid transport and metabolism]. |
| COG0412 | DLH | 1.93e-05 | 522 | 657 | 18 | 141 | Dienelactone hydrolase [Secondary metabolites biosynthesis, transport and catabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QNL48665.1 | 4.44e-310 | 6 | 772 | 8 | 775 |
| QNL48806.1 | 9.04e-296 | 10 | 777 | 5 | 779 |
| AKP51248.1 | 8.01e-224 | 1 | 776 | 1 | 767 |
| AEL25656.1 | 1.37e-218 | 32 | 776 | 32 | 764 |
| BBD44973.1 | 7.57e-207 | 26 | 768 | 45 | 789 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7NN3_A | 9.46e-33 | 94 | 403 | 32 | 365 | ChainA, Beta-xylanase [Caldicellulosiruptor kristjanssonii I77R1B],7NN3_B Chain B, Beta-xylanase [Caldicellulosiruptor kristjanssonii I77R1B],7NN3_C Chain C, Beta-xylanase [Caldicellulosiruptor kristjanssonii I77R1B],7NN3_D Chain D, Beta-xylanase [Caldicellulosiruptor kristjanssonii I77R1B] |
| 6EHN_A | 3.49e-31 | 85 | 401 | 14 | 396 | Structuralinsight into a promiscuous CE15 esterase from the marine bacterial metagenome [unidentified prokaryotic organism] |
| 6GRY_A | 1.81e-24 | 100 | 405 | 26 | 397 | GlucuronoylEsterase from Solibacter usitatus. [Candidatus Solibacter usitatus] |
| 6GRW_A | 3.74e-23 | 229 | 401 | 211 | 394 | GlucuronoylEsterase from Opitutus terrae (Au derivative) [Opitutus terrae PB90-1],6GS0_A Native Glucuronoyl Esterase from Opitutus terrae [Opitutus terrae PB90-1] |
| 6SYU_A | 4.62e-23 | 229 | 401 | 229 | 412 | Thewild type glucuronoyl esterase OtCE15A from Opitutus terrae in complex with xylobiose [Opitutus terrae],6T0I_A The wild type glucuronoyl esterase OtCE15A from Opitutus terrae in complex with the aldotetrauronic acid XUX [Opitutus terrae] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| A0A0K2VM55 | 3.23e-30 | 85 | 401 | 42 | 424 | Carbohydrate esterase MZ0003 OS=Unknown prokaryotic organism OX=2725 GN=MZ0003 PE=1 SV=1 |
| P0CU53 | 9.44e-22 | 85 | 387 | 45 | 352 | 4-O-methyl-glucuronoyl methylesterase 1 OS=Wolfiporia cocos (strain MD-104) OX=742152 GN=WOLCODRAFT_23632 PE=1 SV=1 |
| K5XDZ6 | 2.40e-19 | 85 | 387 | 44 | 350 | 4-O-methyl-glucuronoyl methylesterase OS=Phanerochaete carnosa (strain HHB-10118-sp) OX=650164 GN=PHACADRAFT_247750 PE=1 SV=1 |
| P0CT88 | 1.39e-18 | 100 | 387 | 56 | 350 | 4-O-methyl-glucuronoyl methylesterase 2 OS=Phanerochaete chrysosporium (strain RP-78 / ATCC MYA-4764 / FGSC 9002) OX=273507 GN=e_gw1.11.1537.1 PE=1 SV=1 |
| A0A0A7EQR3 | 2.37e-18 | 85 | 390 | 112 | 422 | 4-O-methyl-glucuronoyl methylesterase OS=Cerrena unicolor OX=90312 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
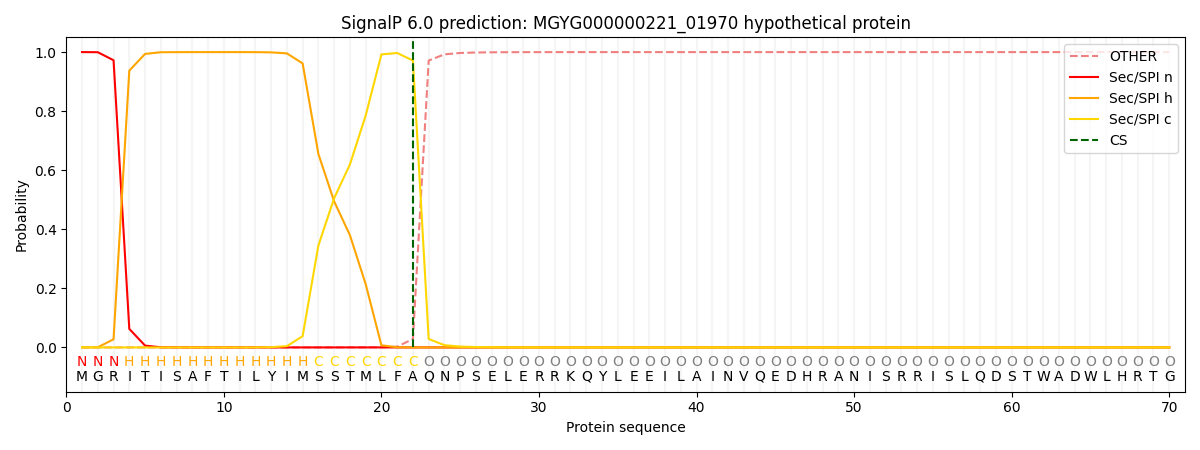
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000324 | 0.999052 | 0.000164 | 0.000166 | 0.000154 | 0.000144 |
