You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000222_00976
Basic Information
help
| Species |
Parabacteroides sp003473295
|
| Lineage |
Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Tannerellaceae; Parabacteroides; Parabacteroides sp003473295
|
| CAZyme ID |
MGYG000000222_00976
|
| CAZy Family |
GH136 |
| CAZyme Description |
hypothetical protein
|
| CAZyme Property |
| Protein Length |
CGC |
Molecular Weight |
Isoelectric Point |
| 503 |
|
56143.52 |
7.0599 |
|
| Genome Property |
| Genome Assembly ID |
Genome Size |
Genome Type |
Country |
Continent |
| MGYG000000222 |
4189286 |
Isolate |
China |
Asia |
|
| Gene Location |
Start: 22186;
End: 23697
Strand: -
|
No EC number prediction in MGYG000000222_00976.
| Family |
Start |
End |
Evalue |
family coverage |
| GH136 |
25 |
442 |
4.7e-96 |
0.8329938900203666 |
MGYG000000222_00976 has no CDD domain.
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
Description |
| 6KQT_A
|
6.48e-28 |
28 |
352 |
247 |
617 |
CrystalStructure of GH136 lacto-N-biosidase from Eubacterium ramulus - native protein [Eubacterium ramulus ATCC 29099] |
| 6KQS_A
|
2.80e-27 |
28 |
352 |
247 |
617 |
CrystalStructure of GH136 lacto-N-biosidase from Eubacterium ramulus - selenomethionine derivative [Eubacterium ramulus ATCC 29099] |
| 7V6M_A
|
2.85e-27 |
23 |
382 |
6 |
426 |
ChainA, Fibronectin type III domain-containing protein [Tyzzerella nexilis] |
This protein is predicted as LIPO
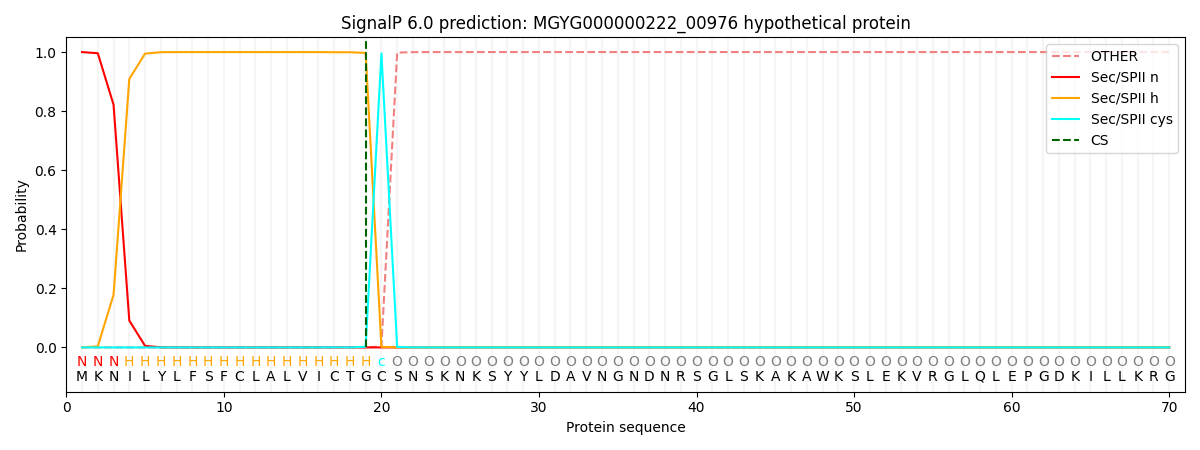
| Other |
SP_Sec_SPI |
LIPO_Sec_SPII |
TAT_Tat_SPI |
TATLIP_Sec_SPII |
PILIN_Sec_SPIII |
|
0.000000
|
0.000000
|
1.000059
|
0.000000
|
0.000000
|
0.000000
|
There is no transmembrane helices in MGYG000000222_00976.

