You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000224_00072
You are here: Home > Sequence: MGYG000000224_00072
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Bacteroides sp003545565 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; Bacteroides sp003545565 | |||||||||||
| CAZyme ID | MGYG000000224_00072 | |||||||||||
| CAZy Family | GH28 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 87730; End: 89124 Strand: - | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG5434 | Pgu1 | 5.46e-12 | 200 | 395 | 230 | 421 | Polygalacturonase [Carbohydrate transport and metabolism]. |
| pfam00295 | Glyco_hydro_28 | 9.19e-10 | 191 | 445 | 88 | 307 | Glycosyl hydrolases family 28. Glycosyl hydrolase family 28 includes polygalacturonase EC:3.2.1.15 as well as rhamnogalacturonase A(RGase A), EC:3.2.1.-. These enzymes are important in cell wall metabolism. |
| PLN03010 | PLN03010 | 4.67e-05 | 171 | 445 | 115 | 381 | polygalacturonase |
| PLN02793 | PLN02793 | 8.39e-05 | 213 | 377 | 229 | 396 | Probable polygalacturonase |
| pfam13229 | Beta_helix | 0.004 | 191 | 306 | 3 | 109 | Right handed beta helix region. This region contains a parallel beta helix region that shares some similarity with Pectate lyases. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ALJ62050.1 | 1.01e-284 | 19 | 464 | 16 | 461 |
| QUU00181.1 | 1.61e-284 | 1 | 464 | 1 | 464 |
| QUT92200.1 | 2.04e-284 | 19 | 464 | 16 | 461 |
| QUT34117.1 | 1.08e-282 | 1 | 464 | 1 | 464 |
| QUT85436.1 | 9.78e-276 | 1 | 464 | 1 | 461 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| B8MW78 | 1.14e-08 | 192 | 443 | 254 | 478 | Probable endopolygalacturonase D OS=Aspergillus flavus (strain ATCC 200026 / FGSC A1120 / IAM 13836 / NRRL 3357 / JCM 12722 / SRRC 167) OX=332952 GN=pgaD PE=3 SV=1 |
| Q2UT29 | 1.14e-08 | 192 | 443 | 254 | 478 | Probable endopolygalacturonase D OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=pgaD PE=3 SV=2 |
| Q7M1E7 | 7.44e-06 | 214 | 330 | 186 | 294 | Polygalacturonase OS=Chamaecyparis obtusa OX=13415 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
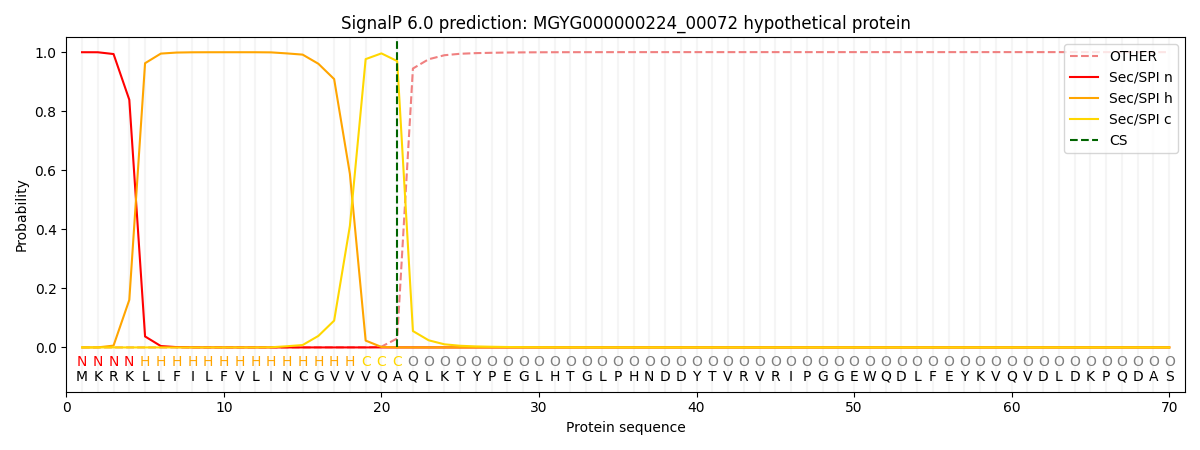
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000479 | 0.997962 | 0.001009 | 0.000168 | 0.000174 | 0.000169 |
