You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000226_00450
You are here: Home > Sequence: MGYG000000226_00450
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
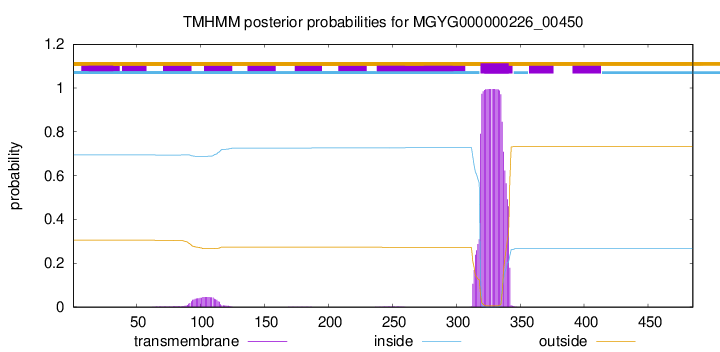
TMHMM annotations
Basic Information help
| Species | Lactococcus lactis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Lactobacillales; Streptococcaceae; Lactococcus; Lactococcus lactis | |||||||||||
| CAZyme ID | MGYG000000226_00450 | |||||||||||
| CAZy Family | CBM50 | |||||||||||
| CAZyme Description | Vitamin B12 import ATP-binding protein BtuD | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 494810; End: 496267 Strand: - | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG1134 | TagH | 3.96e-106 | 14 | 258 | 1 | 249 | ABC-type polysaccharide/polyol phosphate transport system, ATPase component [Carbohydrate transport and metabolism, Cell wall/membrane/envelope biogenesis]. |
| cd03220 | ABC_KpsT_Wzt | 2.85e-96 | 17 | 238 | 1 | 224 | ATP-binding cassette component of polysaccharide transport system. The KpsT/Wzt ABC transporter subfamily is involved in extracellular polysaccharide export. Among the variety of membrane-linked or extracellular polysaccharides excreted by bacteria, only capsular polysaccharides, lipopolysaccharides, and teichoic acids have been shown to be exported by ABC transporters. A typical system is made of a conserved integral membrane and an ABC. In addition to these proteins, capsular polysaccharide exporter systems require two 'accessory' proteins to perform their function: a periplasmic (E.coli) or a lipid-anchored outer membrane protein called OMA (Neisseria meningitidis and Haemophilus influenza) and a cytoplasmic membrane protein MPA2. |
| PRK13545 | tagH | 4.75e-84 | 16 | 322 | 4 | 301 | teichoic acids export protein ATP-binding subunit; Provisional |
| PRK13546 | PRK13546 | 1.05e-77 | 17 | 269 | 5 | 256 | teichoic acids export ABC transporter ATP-binding subunit TagH. |
| COG1131 | CcmA | 1.63e-41 | 34 | 303 | 2 | 285 | ABC-type multidrug transport system, ATPase component [Defense mechanisms]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AIS04007.1 | 0.0 | 7 | 485 | 1 | 479 |
| QQF01236.1 | 0.0 | 7 | 485 | 1 | 479 |
| QEA60341.1 | 0.0 | 7 | 485 | 1 | 479 |
| AII12421.1 | 0.0 | 1 | 485 | 1 | 485 |
| SBW30197.1 | 0.0 | 1 | 485 | 1 | 485 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7DD0_A | 7.33e-83 | 14 | 266 | 2 | 252 | Crystalstructure of the N-terminal domain of TagH from Bacillus subtilis [Bacillus subtilis subsp. subtilis str. 168],7DD0_B Crystal structure of the N-terminal domain of TagH from Bacillus subtilis [Bacillus subtilis subsp. subtilis str. 168],7DD0_C Crystal structure of the N-terminal domain of TagH from Bacillus subtilis [Bacillus subtilis subsp. subtilis str. 168],7DD0_D Crystal structure of the N-terminal domain of TagH from Bacillus subtilis [Bacillus subtilis subsp. subtilis str. 168] |
| 6JBH_A | 4.68e-81 | 21 | 273 | 9 | 260 | Cryo-EMstructure and transport mechanism of a wall teichoic acid ABC transporter [Alicyclobacillus herbarius],6JBH_B Cryo-EM structure and transport mechanism of a wall teichoic acid ABC transporter [Alicyclobacillus herbarius] |
| 6AMX_A | 6.89e-51 | 17 | 247 | 1 | 231 | CrystalStructure of Nucelotide Binding Domain of O-antigen polysaccharide ABC-transporter [Aquifex aeolicus VF5],6AN5_A Crystal Structure of The Nucelotide Binding Domain of an O-antigen polysaccharide ABC-transporter [Aquifex aeolicus VF5],6AN5_B Crystal Structure of The Nucelotide Binding Domain of an O-antigen polysaccharide ABC-transporter [Aquifex aeolicus VF5],6OIH_A Crystal structure of O-antigen polysaccharide ABC-transporter [Aquifex aeolicus VF5],6OIH_B Crystal structure of O-antigen polysaccharide ABC-transporter [Aquifex aeolicus VF5] |
| 6M96_A | 2.01e-50 | 17 | 247 | 3 | 233 | ATP-boundconformation of the WzmWzt O antigen ABC transporter [Aquifex aeolicus VF5] |
| 7K2T_A | 1.28e-48 | 17 | 247 | 3 | 233 | ChainA, ABC transporter [Aquifex aeolicus VF5],7K2T_C Chain C, ABC transporter [Aquifex aeolicus VF5] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q9CH26 | 1.14e-128 | 14 | 483 | 5 | 459 | Teichoic acids export ATP-binding protein TagH OS=Lactococcus lactis subsp. lactis (strain IL1403) OX=272623 GN=tagH PE=3 SV=1 |
| Q831L8 | 6.43e-106 | 13 | 478 | 3 | 444 | Teichoic acids export ATP-binding protein TagH OS=Enterococcus faecalis (strain ATCC 700802 / V583) OX=226185 GN=tagH PE=3 SV=1 |
| Q03SI5 | 1.32e-84 | 16 | 307 | 6 | 297 | Teichoic acids export ATP-binding protein TagH OS=Levilactobacillus brevis (strain ATCC 367 / BCRC 12310 / CIP 105137 / JCM 1170 / LMG 11437 / NCIMB 947 / NCTC 947) OX=387344 GN=tagH PE=3 SV=1 |
| Q65E84 | 2.68e-83 | 14 | 284 | 2 | 266 | Teichoic acids export ATP-binding protein TagH OS=Bacillus licheniformis (strain ATCC 14580 / DSM 13 / JCM 2505 / CCUG 7422 / NBRC 12200 / NCIMB 9375 / NCTC 10341 / NRRL NRS-1264 / Gibson 46) OX=279010 GN=tagH PE=3 SV=1 |
| Q88ZH4 | 2.98e-80 | 16 | 348 | 6 | 342 | Teichoic acids export ATP-binding protein TagH OS=Lactiplantibacillus plantarum (strain ATCC BAA-793 / NCIMB 8826 / WCFS1) OX=220668 GN=tagH PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
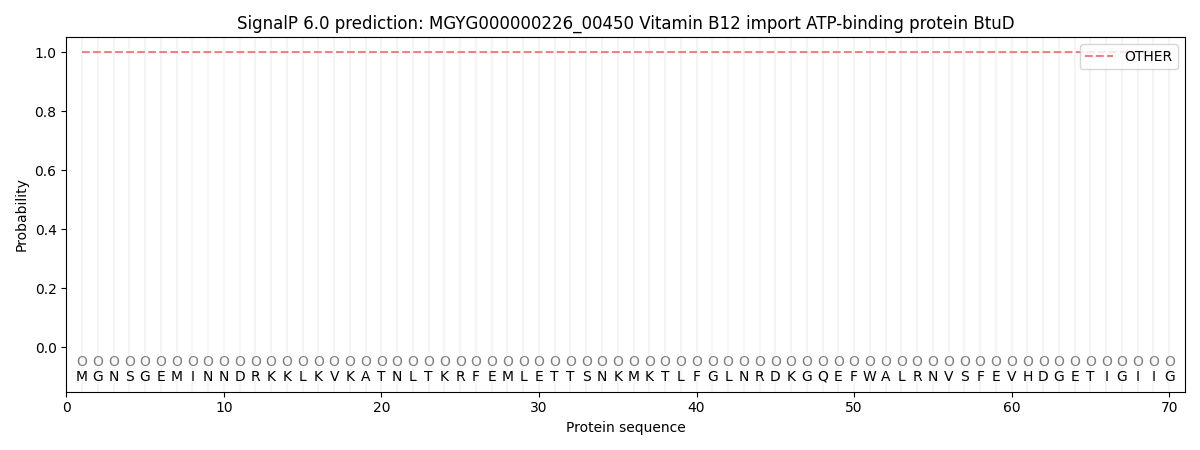
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000071 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |