You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000226_01565
You are here: Home > Sequence: MGYG000000226_01565
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
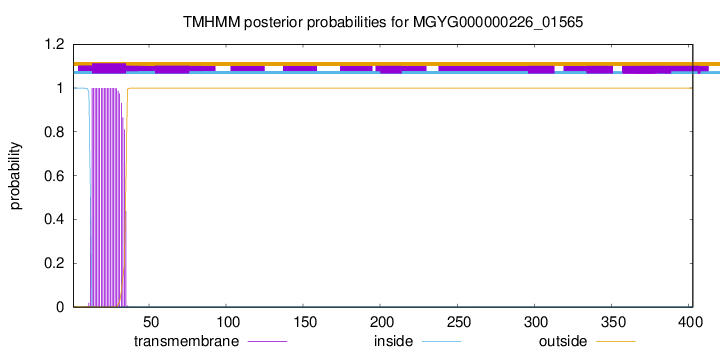
TMHMM annotations
Basic Information help
| Species | Lactococcus lactis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Lactobacillales; Streptococcaceae; Lactococcus; Lactococcus lactis | |||||||||||
| CAZyme ID | MGYG000000226_01565 | |||||||||||
| CAZy Family | GH3 | |||||||||||
| CAZyme Description | Beta-hexosaminidase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 262629; End: 263840 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH3 | 132 | 357 | 2.2e-40 | 0.9537037037037037 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG1472 | BglX | 4.74e-85 | 69 | 403 | 1 | 319 | Periplasmic beta-glucosidase and related glycosidases [Carbohydrate transport and metabolism]. |
| pfam00933 | Glyco_hydro_3 | 6.38e-44 | 70 | 395 | 1 | 316 | Glycosyl hydrolase family 3 N terminal domain. |
| PRK05337 | PRK05337 | 2.97e-38 | 100 | 357 | 25 | 278 | beta-hexosaminidase; Provisional |
| PRK15098 | PRK15098 | 3.31e-08 | 66 | 401 | 42 | 358 | beta-glucosidase BglX. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AIS03052.1 | 1.52e-281 | 1 | 403 | 1 | 403 |
| QQF00548.1 | 1.77e-280 | 1 | 403 | 1 | 403 |
| QEA62019.1 | 7.19e-280 | 1 | 403 | 1 | 403 |
| AUS68974.1 | 1.14e-276 | 1 | 403 | 1 | 403 |
| ARD93016.1 | 1.14e-276 | 1 | 403 | 1 | 403 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5BU9_A | 1.46e-100 | 69 | 396 | 5 | 337 | Crystalstructure of Beta-N-acetylhexosaminidase from Beutenbergia cavernae DSM 12333 [Beutenbergia cavernae DSM 12333],5BU9_B Crystal structure of Beta-N-acetylhexosaminidase from Beutenbergia cavernae DSM 12333 [Beutenbergia cavernae DSM 12333] |
| 6K5J_A | 2.22e-30 | 65 | 397 | 7 | 338 | Structureof a glycoside hydrolase family 3 beta-N-acetylglucosaminidase from Paenibacillus sp. str. FPU-7 [Paenibacillaceae] |
| 2OXN_A | 1.88e-26 | 101 | 357 | 26 | 271 | Vibriocholerae family 3 glycoside hydrolase (NagZ) in complex with PUGNAc [Vibrio cholerae] |
| 1Y65_A | 1.94e-26 | 101 | 357 | 28 | 273 | Crystalstructure of beta-hexosaminidase from Vibrio cholerae in complex with N-acetyl-D-glucosamine to a resolution of 1.85 [Vibrio cholerae] |
| 3GS6_A | 3.55e-26 | 101 | 357 | 26 | 271 | ChainA, Beta-hexosaminidase [Vibrio cholerae],3GSM_A Chain A, Beta-hexosaminidase [Vibrio cholerae] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q1H075 | 1.13e-30 | 101 | 377 | 28 | 302 | Beta-hexosaminidase OS=Methylobacillus flagellatus (strain KT / ATCC 51484 / DSM 6875) OX=265072 GN=nagZ PE=3 SV=1 |
| Q0A911 | 5.39e-29 | 100 | 380 | 27 | 300 | Beta-hexosaminidase OS=Alkalilimnicola ehrlichii (strain ATCC BAA-1101 / DSM 17681 / MLHE-1) OX=187272 GN=nagZ PE=3 SV=1 |
| Q31G32 | 6.29e-29 | 71 | 371 | 1 | 304 | Beta-hexosaminidase OS=Hydrogenovibrio crunogenus (strain DSM 25203 / XCL-2) OX=317025 GN=nagZ PE=3 SV=1 |
| Q0AF74 | 4.82e-27 | 101 | 371 | 28 | 296 | Beta-hexosaminidase OS=Nitrosomonas eutropha (strain DSM 101675 / C91 / Nm57) OX=335283 GN=nagZ PE=3 SV=1 |
| C3LSU7 | 8.71e-26 | 101 | 357 | 26 | 271 | Beta-hexosaminidase OS=Vibrio cholerae serotype O1 (strain M66-2) OX=579112 GN=nagZ PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
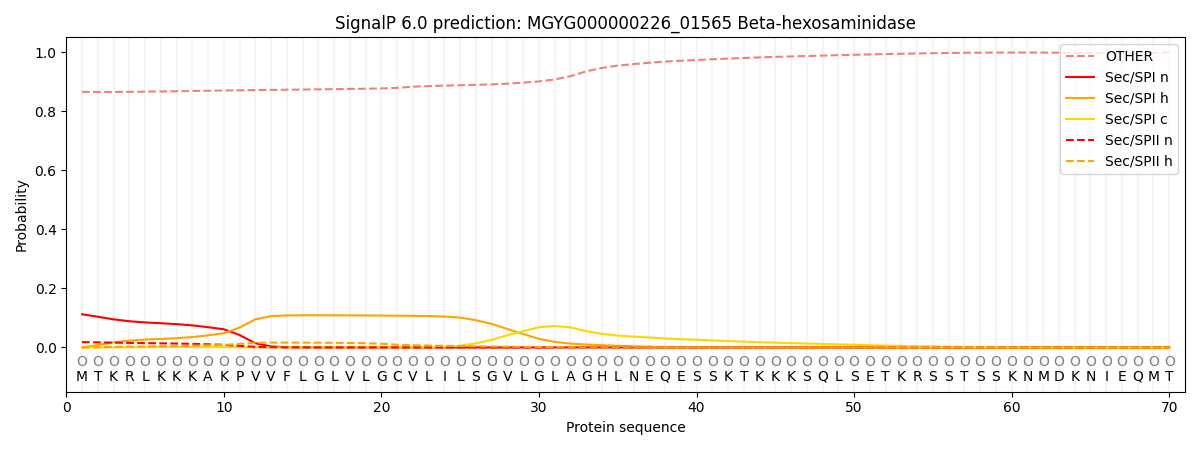
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.870532 | 0.103556 | 0.018856 | 0.000881 | 0.000424 | 0.005764 |