You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000227_02828
You are here: Home > Sequence: MGYG000000227_02828
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Bacillus sonorensis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Bacillales; Bacillaceae; Bacillus; Bacillus sonorensis | |||||||||||
| CAZyme ID | MGYG000000227_02828 | |||||||||||
| CAZy Family | GH126 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 165065; End: 166159 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH126 | 45 | 353 | 9.6e-100 | 0.9875389408099688 |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ASB91428.1 | 9.24e-270 | 1 | 364 | 1 | 364 |
| QAT67608.1 | 1.56e-213 | 1 | 364 | 1 | 364 |
| ATH93813.1 | 6.33e-213 | 1 | 364 | 1 | 364 |
| SCA88439.1 | 5.85e-210 | 7 | 364 | 2 | 359 |
| AZV48940.1 | 4.09e-202 | 1 | 364 | 1 | 364 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6R2M_A | 1.55e-38 | 27 | 338 | 4 | 306 | Crystalstructure of PssZ from Listeria monocytogenes [Listeria monocytogenes],6R2M_B Crystal structure of PssZ from Listeria monocytogenes [Listeria monocytogenes] |
| 3REN_A | 9.59e-24 | 18 | 359 | 11 | 350 | CPF_2247,a novel alpha-amylase from Clostridium perfringens [Clostridium perfringens],3REN_B CPF_2247, a novel alpha-amylase from Clostridium perfringens [Clostridium perfringens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| O31486 | 5.12e-194 | 1 | 364 | 1 | 362 | Putative lipoprotein YdaJ OS=Bacillus subtilis (strain 168) OX=224308 GN=ydaJ PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
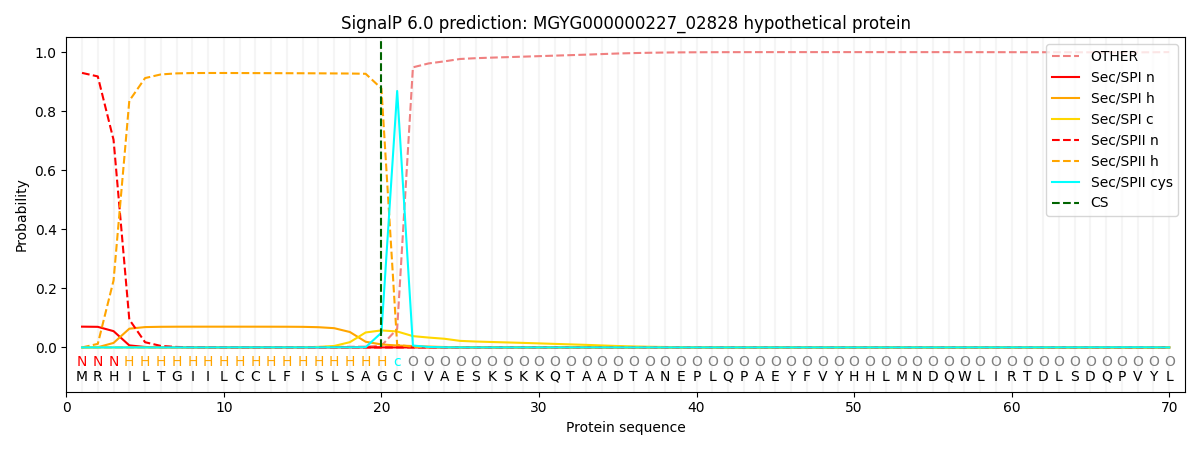
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000391 | 0.067740 | 0.931620 | 0.000091 | 0.000092 | 0.000071 |
