You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000227_03022
You are here: Home > Sequence: MGYG000000227_03022
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Bacillus sonorensis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Bacillales; Bacillaceae; Bacillus; Bacillus sonorensis | |||||||||||
| CAZyme ID | MGYG000000227_03022 | |||||||||||
| CAZy Family | GH3 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 50010; End: 52946 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH3 | 72 | 304 | 7.7e-59 | 0.9722222222222222 |
| CBM6 | 859 | 977 | 2.5e-24 | 0.8768115942028986 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| PRK15098 | PRK15098 | 2.24e-103 | 38 | 839 | 23 | 758 | beta-glucosidase BglX. |
| PLN03080 | PLN03080 | 1.39e-92 | 37 | 801 | 40 | 739 | Probable beta-xylosidase; Provisional |
| COG1472 | BglX | 1.99e-68 | 69 | 439 | 51 | 393 | Periplasmic beta-glucosidase and related glycosidases [Carbohydrate transport and metabolism]. |
| pfam01915 | Glyco_hydro_3_C | 4.10e-51 | 383 | 727 | 1 | 216 | Glycosyl hydrolase family 3 C-terminal domain. This domain is involved in catalysis and may be involved in binding beta-glucan. This domain is found associated with pfam00933. |
| pfam00933 | Glyco_hydro_3 | 1.64e-44 | 75 | 343 | 63 | 316 | Glycosyl hydrolase family 3 N terminal domain. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ASB90778.1 | 0.0 | 1 | 930 | 1 | 930 |
| SCA87757.1 | 0.0 | 1 | 978 | 1 | 977 |
| QAT66941.1 | 0.0 | 1 | 978 | 1 | 977 |
| ATH93133.1 | 0.0 | 1 | 978 | 1 | 977 |
| AOP16766.1 | 0.0 | 1 | 978 | 1 | 981 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5Z87_A | 1.11e-72 | 63 | 839 | 108 | 778 | ChainA, EmGH1 [Aurantiacibacter marinus],5Z87_B Chain B, EmGH1 [Aurantiacibacter marinus] |
| 7MS2_A | 5.98e-72 | 50 | 840 | 5 | 662 | ChainA, Thermostable beta-glucosidase B [Acetivibrio thermocellus AD2],7MS2_B Chain B, Thermostable beta-glucosidase B [Acetivibrio thermocellus AD2] |
| 3AC0_A | 9.44e-67 | 50 | 849 | 6 | 839 | Crystalstructure of Beta-glucosidase from Kluyveromyces marxianus in complex with glucose [Kluyveromyces marxianus],3AC0_B Crystal structure of Beta-glucosidase from Kluyveromyces marxianus in complex with glucose [Kluyveromyces marxianus],3AC0_C Crystal structure of Beta-glucosidase from Kluyveromyces marxianus in complex with glucose [Kluyveromyces marxianus],3AC0_D Crystal structure of Beta-glucosidase from Kluyveromyces marxianus in complex with glucose [Kluyveromyces marxianus] |
| 4I3G_A | 2.60e-66 | 49 | 839 | 53 | 821 | CrystalStructure of DesR, a beta-glucosidase from Streptomyces venezuelae in complex with D-glucose. [Streptomyces venezuelae],4I3G_B Crystal Structure of DesR, a beta-glucosidase from Streptomyces venezuelae in complex with D-glucose. [Streptomyces venezuelae] |
| 5TF0_A | 2.79e-66 | 75 | 839 | 80 | 744 | CrystalStructure of Glycosil Hydrolase Family 3 N-Terminal Domain Protein from Bacteroides intestinalis [Bacteroides intestinalis DSM 17393],5TF0_B Crystal Structure of Glycosil Hydrolase Family 3 N-Terminal Domain Protein from Bacteroides intestinalis [Bacteroides intestinalis DSM 17393] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| D5EY15 | 1.67e-98 | 32 | 842 | 17 | 856 | Xylan 1,4-beta-xylosidase OS=Prevotella ruminicola (strain ATCC 19189 / JCM 8958 / 23) OX=264731 GN=xyl3A PE=1 SV=1 |
| T2KMH0 | 1.46e-97 | 41 | 841 | 28 | 718 | Beta-xylosidase OS=Formosa agariphila (strain DSM 15362 / KCTC 12365 / LMG 23005 / KMM 3901 / M-2Alg 35-1) OX=1347342 GN=BN863_22130 PE=1 SV=1 |
| Q94KD8 | 6.49e-87 | 46 | 840 | 51 | 758 | Probable beta-D-xylosidase 2 OS=Arabidopsis thaliana OX=3702 GN=BXL2 PE=2 SV=1 |
| P27034 | 2.21e-79 | 50 | 839 | 2 | 803 | Beta-glucosidase OS=Rhizobium radiobacter OX=358 GN=cbg-1 PE=3 SV=1 |
| Q9SGZ5 | 2.08e-78 | 37 | 834 | 36 | 759 | Probable beta-D-xylosidase 7 OS=Arabidopsis thaliana OX=3702 GN=BXL7 PE=2 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
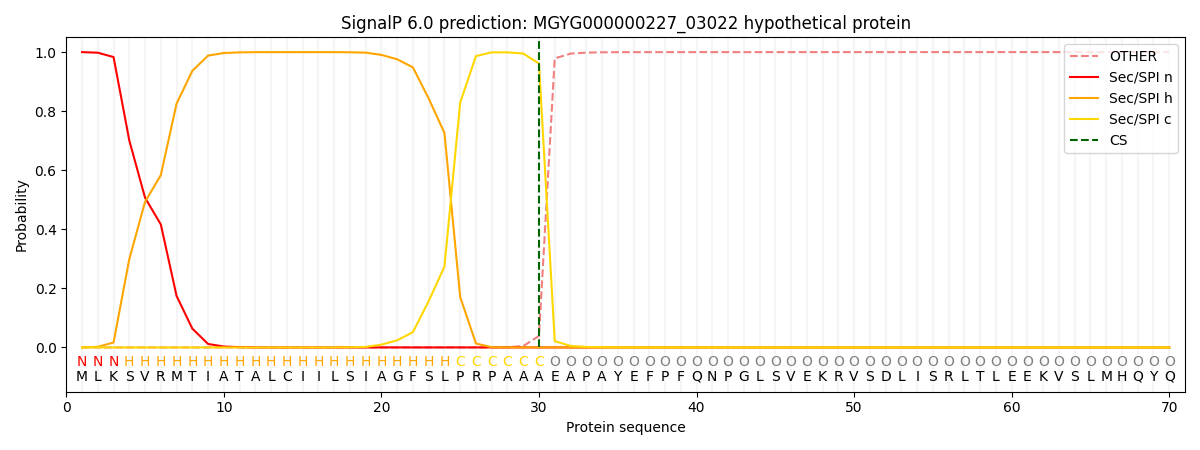
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000391 | 0.998762 | 0.000306 | 0.000188 | 0.000166 | 0.000144 |
