You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000236_03294
You are here: Home > Sequence: MGYG000000236_03294
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Bacteroides fragilis_A | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; Bacteroides fragilis_A | |||||||||||
| CAZyme ID | MGYG000000236_03294 | |||||||||||
| CAZy Family | CBM32 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 94954; End: 95907 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM32 | 187 | 314 | 7.1e-20 | 0.9435483870967742 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam08522 | DUF1735 | 9.80e-20 | 29 | 152 | 4 | 120 | Domain of unknown function (DUF1735). This domain of unknown function is found in a number of bacterial proteins including acylhydrolases. The structure of this domain has a beta-sandwich fold. |
| pfam00754 | F5_F8_type_C | 2.02e-16 | 185 | 306 | 1 | 118 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| cd00057 | FA58C | 9.57e-07 | 182 | 307 | 12 | 135 | Substituted updates: Jan 31, 2002 |
| smart00231 | FA58C | 1.74e-05 | 173 | 307 | 3 | 130 | Coagulation factor 5/8 C-terminal domain, discoidin domain. Cell surface-attached carbohydrate-binding domain, present in eukaryotes and assumed to have horizontally transferred to eubacterial genomes. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QRM69709.1 | 7.25e-241 | 1 | 317 | 1 | 317 |
| QCQ53710.1 | 8.47e-240 | 1 | 317 | 1 | 317 |
| QTO27246.1 | 3.45e-239 | 1 | 317 | 1 | 317 |
| QCQ44749.1 | 4.90e-239 | 1 | 317 | 1 | 317 |
| QCQ35820.1 | 4.90e-239 | 1 | 317 | 1 | 317 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3F2Z_A | 1.13e-10 | 176 | 316 | 5 | 145 | Crystalstructure of the C-terminal domain of a chitobiase (BF3579) from Bacteroides fragilis, Northeast Structural Genomics Consortium Target BfR260B [Bacteroides fragilis NCTC 9343] |
| 1GOF_A | 3.94e-09 | 176 | 317 | 9 | 146 | NOVELTHIOETHER BOND REVEALED BY A 1.7 ANGSTROMS CRYSTAL STRUCTURE OF GALACTOSE OXIDASE [Hypomyces rosellus],1GOG_A NOVEL THIOETHER BOND REVEALED BY A 1.7 ANGSTROMS CRYSTAL STRUCTURE OF GALACTOSE OXIDASE [Hypomyces rosellus],1GOH_A NOVEL THIOETHER BOND REVEALED BY A 1.7 ANGSTROMS CRYSTAL STRUCTURE OF GALACTOSE OXIDASE [Hypomyces rosellus],2EIE_A Chain A, Galactose oxidase [Fusarium graminearum],2JKX_A Chain A, GALACTOSE OXIDASE [Fusarium graminearum],2VZ1_A Chain A, GALACTOSE OXIDASE [Fusarium graminearum],2VZ3_A Chain A, Galactose Oxidase [Fusarium graminearum] |
| 1T2X_A | 3.94e-09 | 176 | 317 | 9 | 146 | Glactoseoxidase C383S mutant identified by directed evolution [Fusarium sp.] |
| 2EIB_A | 3.94e-09 | 176 | 317 | 9 | 146 | ChainA, Galactose oxidase [Fusarium graminearum] |
| 2EIC_A | 3.94e-09 | 176 | 317 | 9 | 146 | ChainA, Galactose oxidase [Fusarium graminearum] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P0CS93 | 2.21e-08 | 176 | 317 | 50 | 187 | Galactose oxidase OS=Gibberella zeae OX=5518 GN=GAOA PE=1 SV=1 |
| I1S2N3 | 2.95e-08 | 176 | 317 | 50 | 187 | Galactose oxidase OS=Gibberella zeae (strain ATCC MYA-4620 / CBS 123657 / FGSC 9075 / NRRL 31084 / PH-1) OX=229533 GN=GAOA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
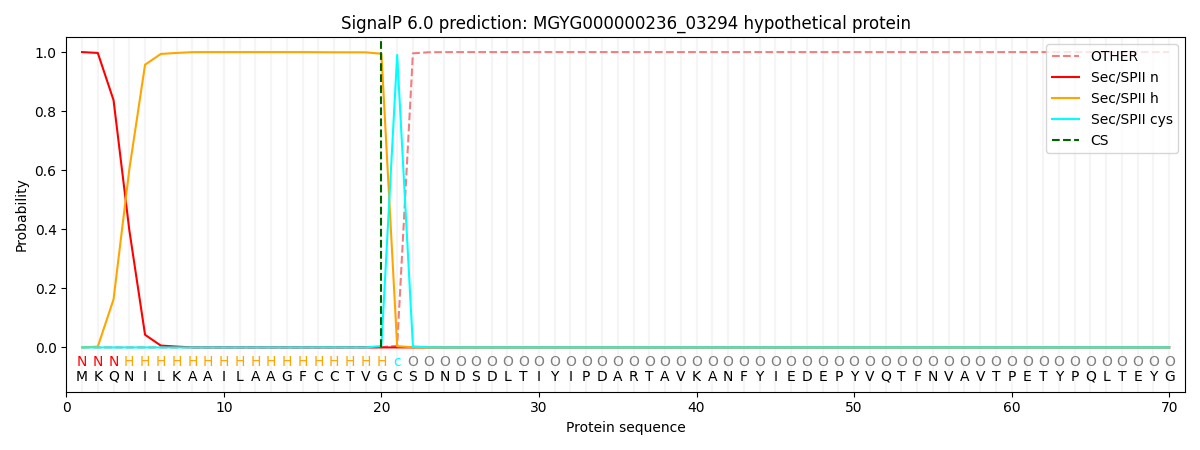
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000001 | 1.000032 | 0.000000 | 0.000000 | 0.000000 |
