You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000238_02685
You are here: Home > Sequence: MGYG000000238_02685
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Enterococcus_D casseliflavus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Lactobacillales; Enterococcaceae; Enterococcus_D; Enterococcus_D casseliflavus | |||||||||||
| CAZyme ID | MGYG000000238_02685 | |||||||||||
| CAZy Family | GT2 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 27144; End: 27974 Strand: + | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd02526 | GT2_RfbF_like | 3.99e-13 | 26 | 206 | 18 | 206 | RfbF is a putative dTDP-rhamnosyl transferase. Shigella flexneri RfbF protein is a putative dTDP-rhamnosyl transferase. dTDP rhamnosyl transferases of Shigella flexneri add rhamnose sugars to N-acetyl-glucosamine in the O-antigen tetrasaccharide repeat. Lipopolysaccharide O antigens are important virulence determinants for many bacteria. The variations of sugar composition, the sequence of the sugars and the linkages in the O antigen provide structural diversity of the O antigen. |
| COG1216 | GT2 | 4.39e-04 | 1 | 204 | 2 | 220 | Glycosyltransferase, GT2 family [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QOG31433.1 | 8.44e-199 | 1 | 276 | 1 | 276 |
| AYJ45360.1 | 8.10e-197 | 1 | 276 | 1 | 276 |
| QGN30130.1 | 1.63e-196 | 1 | 276 | 1 | 276 |
| AMG50185.1 | 2.71e-195 | 1 | 276 | 1 | 276 |
| ALS37340.1 | 5.55e-68 | 2 | 268 | 10 | 277 |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
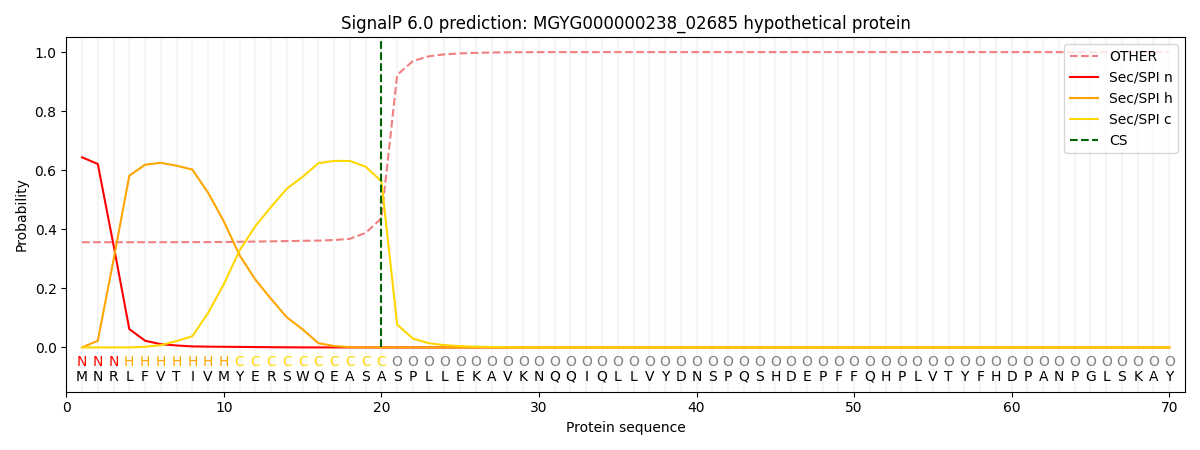
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.359617 | 0.639365 | 0.000259 | 0.000284 | 0.000221 | 0.000221 |
