You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000243_00810
You are here: Home > Sequence: MGYG000000243_00810
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Phocaeicola massiliensis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Phocaeicola; Phocaeicola massiliensis | |||||||||||
| CAZyme ID | MGYG000000243_00810 | |||||||||||
| CAZy Family | CBM62 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 125147; End: 127075 Strand: + | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| smart00460 | TGc | 4.60e-04 | 217 | 273 | 1 | 65 | Transglutaminase/protease-like homologues. Transglutaminases are enzymes that establish covalent links between proteins. A subset of transglutaminase homologues appear to catalyse the reverse reaction, the hydrolysis of peptide bonds. Proteins with this domain are both extracellular and intracellular, and it is likely that the eukaryotic intracellular proteins are involved in signalling events. |
| pfam00754 | F5_F8_type_C | 0.009 | 515 | 613 | 2 | 105 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QCQ52208.1 | 1.18e-218 | 10 | 642 | 20 | 653 |
| QCQ55964.1 | 1.51e-218 | 4 | 642 | 10 | 650 |
| QTO25417.1 | 2.14e-218 | 4 | 642 | 10 | 650 |
| QCQ39038.1 | 4.77e-218 | 10 | 642 | 20 | 653 |
| QCQ33598.1 | 8.64e-218 | 4 | 642 | 10 | 650 |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as LIPO
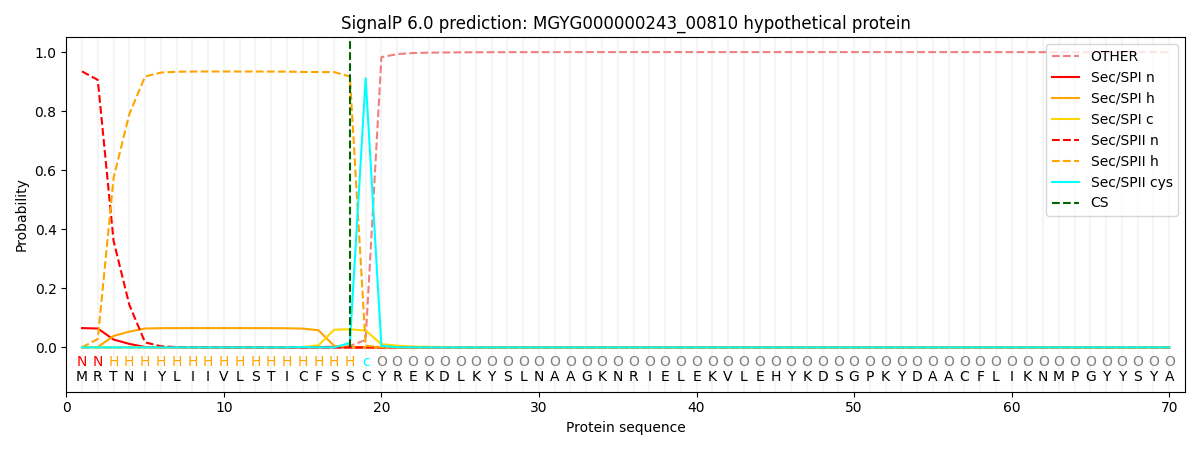
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000150 | 0.063536 | 0.936242 | 0.000027 | 0.000039 | 0.000028 |
