You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000247_00632
You are here: Home > Sequence: MGYG000000247_00632
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Collinsella intestinalis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Actinobacteriota; Coriobacteriia; Coriobacteriales; Coriobacteriaceae; Collinsella; Collinsella intestinalis | |||||||||||
| CAZyme ID | MGYG000000247_00632 | |||||||||||
| CAZy Family | GH73 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 735639; End: 736694 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH73 | 222 | 346 | 8.7e-19 | 0.9609375 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam18885 | DUF5648 | 2.15e-14 | 85 | 122 | 1 | 39 | Repeat of unknown function (DUF5648). This entry represents a repeat of approximately 40 residues in length. It is often associated with enzymatic domains in bacterial cell surface proteins. This entry may represent a beta-propeller repeat, although most proteins only possess three repeats rather than the expected 6-8 copies. |
| pfam18885 | DUF5648 | 1.53e-12 | 130 | 167 | 1 | 39 | Repeat of unknown function (DUF5648). This entry represents a repeat of approximately 40 residues in length. It is often associated with enzymatic domains in bacterial cell surface proteins. This entry may represent a beta-propeller repeat, although most proteins only possess three repeats rather than the expected 6-8 copies. |
| pfam18885 | DUF5648 | 6.98e-10 | 39 | 76 | 1 | 39 | Repeat of unknown function (DUF5648). This entry represents a repeat of approximately 40 residues in length. It is often associated with enzymatic domains in bacterial cell surface proteins. This entry may represent a beta-propeller repeat, although most proteins only possess three repeats rather than the expected 6-8 copies. |
| pfam01832 | Glucosaminidase | 4.36e-09 | 221 | 288 | 1 | 77 | Mannosyl-glycoprotein endo-beta-N-acetylglucosaminidase. This family includes Mannosyl-glycoprotein endo-beta-N-acetylglucosaminidase EC:3.2.1.96. As well as the flageller protein J that has been shown to hydrolyze peptidoglycan. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QTU85010.1 | 2.32e-56 | 176 | 348 | 836 | 1008 |
| QOY61400.1 | 6.23e-53 | 176 | 348 | 12 | 186 |
| CBL26238.1 | 3.72e-44 | 180 | 346 | 84 | 253 |
| QEI32622.1 | 5.82e-44 | 178 | 346 | 1136 | 1309 |
| QRT31212.1 | 5.82e-44 | 178 | 346 | 1136 | 1309 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| O51481 | 1.02e-23 | 225 | 348 | 76 | 197 | Uncharacterized protein BB_0531 OS=Borreliella burgdorferi (strain ATCC 35210 / DSM 4680 / CIP 102532 / B31) OX=224326 GN=BB_0531 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
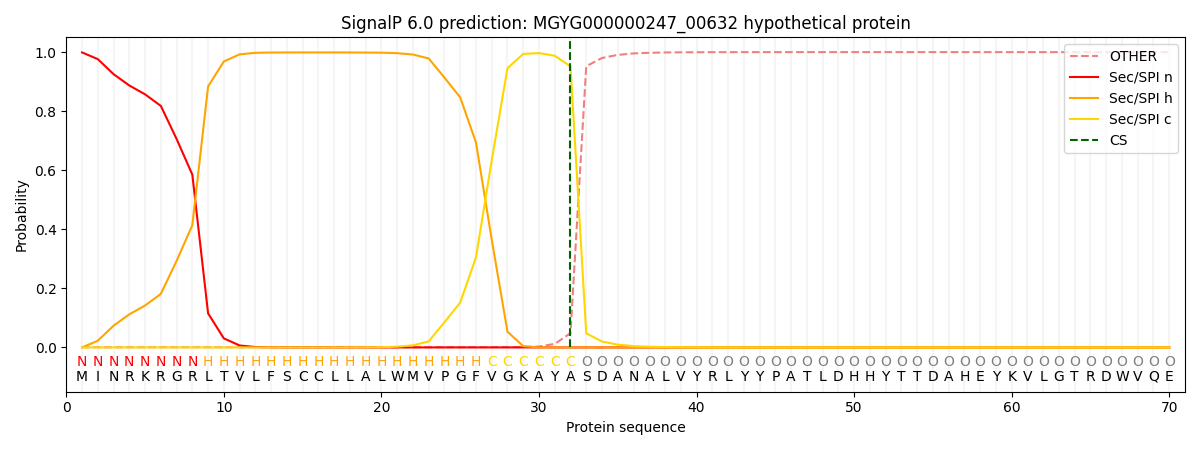
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.001962 | 0.996105 | 0.000493 | 0.000864 | 0.000300 | 0.000246 |

