You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000255_00813
You are here: Home > Sequence: MGYG000000255_00813
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | CAG-81 sp900066055 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; CAG-81; CAG-81 sp900066055 | |||||||||||
| CAZyme ID | MGYG000000255_00813 | |||||||||||
| CAZy Family | GH73 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 97428; End: 99623 Strand: + | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG4193 | LytD | 7.93e-19 | 384 | 535 | 104 | 245 | Beta- N-acetylglucosaminidase [Carbohydrate transport and metabolism]. |
| pfam08239 | SH3_3 | 5.91e-09 | 47 | 107 | 1 | 54 | Bacterial SH3 domain. |
| smart00287 | SH3b | 7.07e-07 | 41 | 107 | 3 | 62 | Bacterial SH3 domain homologues. |
| NF033849 | ser_rich_anae_1 | 2.78e-05 | 264 | 414 | 354 | 499 | serine-rich protein. This serine-rich protein belongs to a family with large size (over 1000 amino acids), which a highly serine-rich central region that averages over 300 aa in length. Species encoding members of this family of proteins tend to be anaerobic bacteria, including Gram-positive bacteria of the human gut microbiome and Chloroflexi from marine sediments. |
| COG4991 | YraI | 0.002 | 25 | 124 | 20 | 112 | Uncharacterized conserved protein YraI [Function unknown]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QJU23484.1 | 4.58e-196 | 3 | 638 | 4 | 686 |
| QIX93811.1 | 1.00e-195 | 3 | 638 | 4 | 680 |
| ANU49989.1 | 3.83e-195 | 3 | 638 | 4 | 680 |
| QQR01104.1 | 3.83e-195 | 3 | 638 | 4 | 680 |
| ASN95383.1 | 1.42e-194 | 3 | 638 | 4 | 686 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6FXO_A | 1.15e-12 | 385 | 534 | 97 | 243 | ChainA, Bifunctional autolysin [Staphylococcus aureus subsp. aureus Mu50] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q8CPQ1 | 8.26e-12 | 385 | 534 | 1188 | 1334 | Bifunctional autolysin OS=Staphylococcus epidermidis (strain ATCC 12228 / FDA PCI 1200) OX=176280 GN=atl PE=3 SV=1 |
| O33635 | 1.87e-11 | 385 | 534 | 1188 | 1334 | Bifunctional autolysin OS=Staphylococcus epidermidis OX=1282 GN=atl PE=1 SV=1 |
| Q5HQB9 | 1.87e-11 | 385 | 534 | 1188 | 1334 | Bifunctional autolysin OS=Staphylococcus epidermidis (strain ATCC 35984 / RP62A) OX=176279 GN=atl PE=3 SV=1 |
| Q99V41 | 7.20e-11 | 385 | 534 | 1101 | 1247 | Bifunctional autolysin OS=Staphylococcus aureus (strain N315) OX=158879 GN=atl PE=1 SV=1 |
| Q931U5 | 7.20e-11 | 385 | 534 | 1101 | 1247 | Bifunctional autolysin OS=Staphylococcus aureus (strain Mu50 / ATCC 700699) OX=158878 GN=atl PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
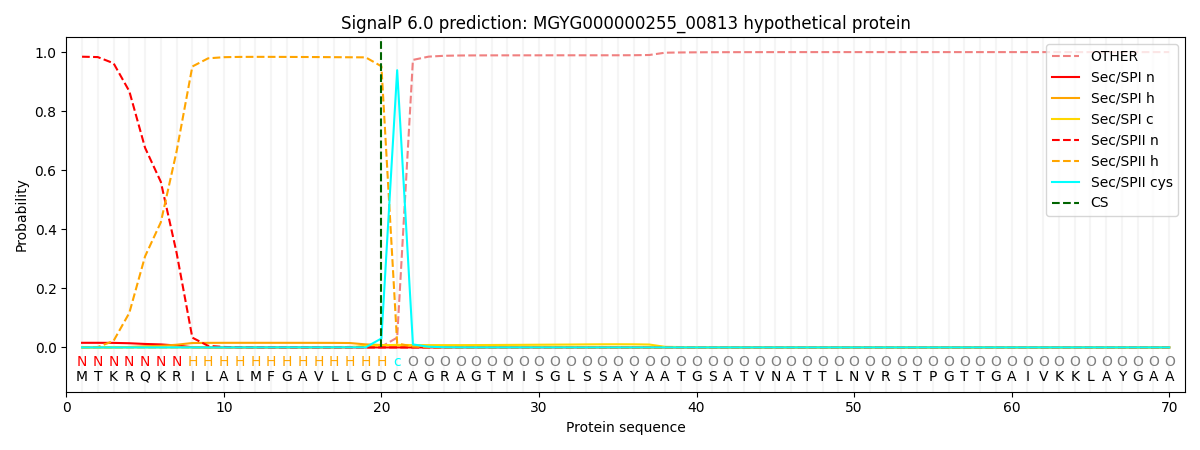
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000019 | 0.015313 | 0.984659 | 0.000007 | 0.000009 | 0.000005 |
