You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000265_00570
You are here: Home > Sequence: MGYG000000265_00570
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Bacteroides nordii | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; Bacteroides nordii | |||||||||||
| CAZyme ID | MGYG000000265_00570 | |||||||||||
| CAZy Family | PL29 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 42248; End: 44101 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| PL29 | 82 | 353 | 4.4e-23 | 0.8803986710963455 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| TIGR04183 | Por_Secre_tail | 7.26e-06 | 557 | 617 | 13 | 72 | Por secretion system C-terminal sorting domain. Species that include Porphyromonas gingivalis, Fibrobacter succinogenes, Flavobacterium johnsoniae, Cytophaga hutchinsonii, Gramella forsetii, Prevotella intermedia, and Salinibacter ruber average twenty or more copies of a C-terminal domain, represented by this model, associated with sorting to the outer membrane and covalent modification. |
| pfam12708 | Pectate_lyase_3 | 2.88e-04 | 81 | 143 | 11 | 67 | Pectate lyase superfamily protein. This family of proteins possesses a beta helical structure like Pectate lyase. This family is most closely related to glycosyl hydrolase family 28. |
| COG4677 | PemB | 0.005 | 12 | 207 | 9 | 221 | Pectin methylesterase and related acyl-CoA thioesterases [Carbohydrate transport and metabolism, Lipid transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AFN75555.1 | 1.08e-119 | 26 | 532 | 25 | 526 |
| ABX04833.1 | 3.42e-96 | 26 | 532 | 309 | 787 |
| ARU26836.1 | 1.38e-86 | 8 | 532 | 609 | 1111 |
| QEC44668.1 | 3.50e-59 | 51 | 538 | 55 | 502 |
| QIH35080.1 | 4.68e-50 | 28 | 532 | 419 | 894 |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
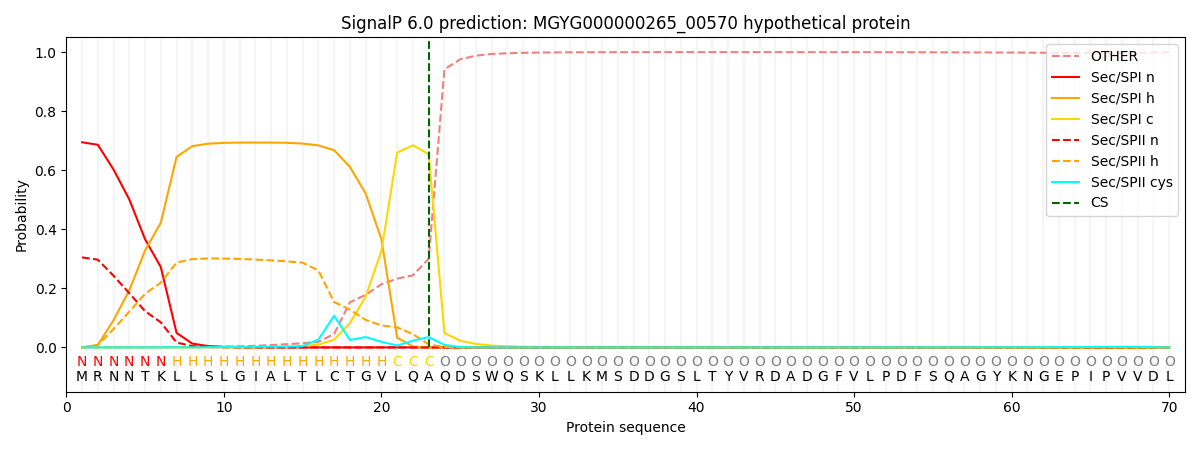
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.001593 | 0.680230 | 0.317046 | 0.000481 | 0.000327 | 0.000295 |
