You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000265_01052
You are here: Home > Sequence: MGYG000000265_01052
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Bacteroides nordii | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; Bacteroides nordii | |||||||||||
| CAZyme ID | MGYG000000265_01052 | |||||||||||
| CAZy Family | GH3 | |||||||||||
| CAZyme Description | Beta-glucosidase BoGH3B | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 179477; End: 181789 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH3 | 120 | 348 | 5.2e-76 | 0.9814814814814815 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| PRK15098 | PRK15098 | 1.28e-177 | 3 | 750 | 7 | 751 | beta-glucosidase BglX. |
| COG1472 | BglX | 1.61e-98 | 44 | 459 | 1 | 377 | Periplasmic beta-glucosidase and related glycosidases [Carbohydrate transport and metabolism]. |
| PLN03080 | PLN03080 | 6.74e-82 | 139 | 750 | 106 | 769 | Probable beta-xylosidase; Provisional |
| pfam00933 | Glyco_hydro_3 | 2.57e-75 | 45 | 381 | 1 | 316 | Glycosyl hydrolase family 3 N terminal domain. |
| pfam01915 | Glyco_hydro_3_C | 2.46e-48 | 418 | 647 | 1 | 216 | Glycosyl hydrolase family 3 C-terminal domain. This domain is involved in catalysis and may be involved in binding beta-glucan. This domain is found associated with pfam00933. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QUT73851.1 | 0.0 | 1 | 768 | 1 | 769 |
| QUT43879.1 | 0.0 | 1 | 765 | 8 | 767 |
| QUT22695.1 | 0.0 | 1 | 766 | 11 | 772 |
| QNL37131.1 | 3.42e-306 | 24 | 765 | 27 | 770 |
| QDH55225.1 | 1.38e-305 | 24 | 765 | 27 | 770 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5Z9S_A | 5.27e-149 | 15 | 765 | 6 | 787 | Functionaland Structural Characterization of a beta-Glucosidase Involved in Saponin Metabolism from Intestinal Bacteria [Bifidobacterium longum],5Z9S_B Functional and Structural Characterization of a beta-Glucosidase Involved in Saponin Metabolism from Intestinal Bacteria [Bifidobacterium longum] |
| 5XXL_A | 5.59e-141 | 33 | 753 | 10 | 741 | Crystalstructure of GH3 beta-glucosidase from Bacteroides thetaiotaomicron [Bacteroides thetaiotaomicron VPI-5482],5XXL_B Crystal structure of GH3 beta-glucosidase from Bacteroides thetaiotaomicron [Bacteroides thetaiotaomicron VPI-5482],5XXM_A Crystal structure of GH3 beta-glucosidase from Bacteroides thetaiotaomicron in complex with gluconolactone [Bacteroides thetaiotaomicron VPI-5482],5XXM_B Crystal structure of GH3 beta-glucosidase from Bacteroides thetaiotaomicron in complex with gluconolactone [Bacteroides thetaiotaomicron VPI-5482] |
| 5XXN_A | 3.09e-140 | 33 | 753 | 10 | 741 | CrystalStructure of mutant (D286N) beta-glucosidase from Bacteroides thetaiotaomicron in complex with sophorose [Bacteroides thetaiotaomicron VPI-5482],5XXN_B Crystal Structure of mutant (D286N) beta-glucosidase from Bacteroides thetaiotaomicron in complex with sophorose [Bacteroides thetaiotaomicron VPI-5482],5XXO_A Crystal structure of mutant (D286N) GH3 beta-glucosidase from Bacteroides thetaiotaomicron in complex with sophorotriose [Bacteroides thetaiotaomicron VPI-5482],5XXO_B Crystal structure of mutant (D286N) GH3 beta-glucosidase from Bacteroides thetaiotaomicron in complex with sophorotriose [Bacteroides thetaiotaomicron VPI-5482] |
| 5TF0_A | 2.62e-139 | 37 | 753 | 13 | 740 | CrystalStructure of Glycosil Hydrolase Family 3 N-Terminal Domain Protein from Bacteroides intestinalis [Bacteroides intestinalis DSM 17393],5TF0_B Crystal Structure of Glycosil Hydrolase Family 3 N-Terminal Domain Protein from Bacteroides intestinalis [Bacteroides intestinalis DSM 17393] |
| 6R5I_A | 1.13e-131 | 34 | 750 | 4 | 719 | ChainA, Periplasmic beta-glucosidase [Pseudomonas aeruginosa PAO1],6R5I_B Chain B, Periplasmic beta-glucosidase [Pseudomonas aeruginosa PAO1],6R5N_A Chain A, Periplasmic beta-glucosidase [Pseudomonas aeruginosa PAO1],6R5N_B Chain B, Periplasmic beta-glucosidase [Pseudomonas aeruginosa PAO1] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| T2KMH0 | 1.26e-149 | 100 | 765 | 37 | 722 | Beta-xylosidase OS=Formosa agariphila (strain DSM 15362 / KCTC 12365 / LMG 23005 / KMM 3901 / M-2Alg 35-1) OX=1347342 GN=BN863_22130 PE=1 SV=1 |
| T2KMH9 | 3.87e-132 | 26 | 753 | 29 | 744 | Putative beta-xylosidase OS=Formosa agariphila (strain DSM 15362 / KCTC 12365 / LMG 23005 / KMM 3901 / M-2Alg 35-1) OX=1347342 GN=BN863_22230 PE=2 SV=1 |
| Q56078 | 9.76e-132 | 37 | 766 | 38 | 765 | Periplasmic beta-glucosidase OS=Salmonella typhimurium (strain LT2 / SGSC1412 / ATCC 700720) OX=99287 GN=bglX PE=3 SV=2 |
| P33363 | 2.64e-127 | 37 | 766 | 38 | 765 | Periplasmic beta-glucosidase OS=Escherichia coli (strain K12) OX=83333 GN=bglX PE=3 SV=2 |
| Q23892 | 4.18e-116 | 37 | 750 | 80 | 812 | Lysosomal beta glucosidase OS=Dictyostelium discoideum OX=44689 GN=gluA PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
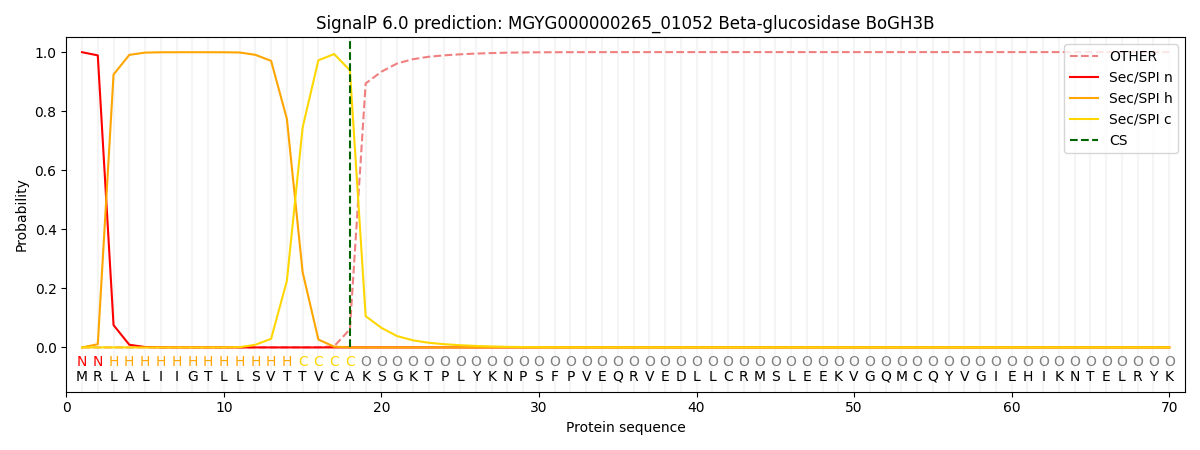
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000550 | 0.998184 | 0.000741 | 0.000187 | 0.000163 | 0.000156 |
