You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000265_01935
You are here: Home > Sequence: MGYG000000265_01935
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Bacteroides nordii | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; Bacteroides nordii | |||||||||||
| CAZyme ID | MGYG000000265_01935 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 63185; End: 64195 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 42 | 317 | 3e-118 | 0.996415770609319 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00150 | Cellulase | 2.70e-29 | 80 | 308 | 36 | 270 | Cellulase (glycosyl hydrolase family 5). |
| COG2730 | BglC | 2.54e-17 | 73 | 310 | 78 | 363 | Aryl-phospho-beta-D-glucosidase BglC, GH1 family [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BCI63367.1 | 5.57e-160 | 1 | 334 | 1 | 332 |
| AHM60064.1 | 5.72e-107 | 23 | 334 | 6 | 320 |
| AXP79804.1 | 3.11e-106 | 35 | 334 | 29 | 331 |
| QMU30925.1 | 3.54e-106 | 1 | 334 | 1 | 335 |
| QJW90914.1 | 6.24e-106 | 35 | 334 | 28 | 330 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5A8N_A | 1.42e-94 | 27 | 331 | 43 | 357 | Crystalstructure of the native form of beta-glucanase SdGluc5_26A from Saccharophagus degradans [Saccharophagus degradans 2-40] |
| 4HTY_A | 2.36e-94 | 35 | 331 | 52 | 352 | CrystalStructure of a metagenome-derived cellulase Cel5A [uncultured bacterium],4HU0_A Crystal Structure of a metagenome-derived cellulase Cel5A in complex with cellotetraose [uncultured bacterium] |
| 5A8O_A | 4.03e-94 | 27 | 331 | 43 | 357 | Crystalstructure of beta-glucanase SdGluc5_26A from Saccharophagus degradans in complex with cellotetraose [Saccharophagus degradans 2-40],5A8P_A Crystal structure beta-glucanase SdGluc5_26A from Saccharophagus degradans in complex with tetrasaccharide B [Saccharophagus degradans 2-40],5A8Q_A Crystal structure beta-glucanase SdGluc5_26A from Saccharophagus degradans in complex with tetrasaccharide A obtained by soaking [Saccharophagus degradans 2-40],5A94_A Crystal structure of beta-Glucanase SdGluc5_26A from Saccharophagus degradans in complex with tetrasaccharide A, form 1 [Saccharophagus degradans 2-40],5A94_B Crystal structure of beta-Glucanase SdGluc5_26A from Saccharophagus degradans in complex with tetrasaccharide A, form 1 [Saccharophagus degradans 2-40],5A94_C Crystal structure of beta-Glucanase SdGluc5_26A from Saccharophagus degradans in complex with tetrasaccharide A, form 1 [Saccharophagus degradans 2-40],5A94_D Crystal structure of beta-Glucanase SdGluc5_26A from Saccharophagus degradans in complex with tetrasaccharide A, form 1 [Saccharophagus degradans 2-40],5A94_E Crystal structure of beta-Glucanase SdGluc5_26A from Saccharophagus degradans in complex with tetrasaccharide A, form 1 [Saccharophagus degradans 2-40],5A94_F Crystal structure of beta-Glucanase SdGluc5_26A from Saccharophagus degradans in complex with tetrasaccharide A, form 1 [Saccharophagus degradans 2-40],5A95_A Crystal structure of beta-glucanase SdGluc5_26A from Saccharophagus degradans in complex with tetrasaccharide A, form 2 [Saccharophagus degradans 2-40],5A95_B Crystal structure of beta-glucanase SdGluc5_26A from Saccharophagus degradans in complex with tetrasaccharide A, form 2 [Saccharophagus degradans 2-40],5A95_C Crystal structure of beta-glucanase SdGluc5_26A from Saccharophagus degradans in complex with tetrasaccharide A, form 2 [Saccharophagus degradans 2-40] |
| 5A8M_A | 1.14e-93 | 27 | 331 | 43 | 357 | Crystalstructure of the selenomethionine derivative of beta-glucanase SdGluc5_26A from Saccharophagus degradans [Saccharophagus degradans 2-40],5A8M_B Crystal structure of the selenomethionine derivative of beta-glucanase SdGluc5_26A from Saccharophagus degradans [Saccharophagus degradans 2-40],5A8M_C Crystal structure of the selenomethionine derivative of beta-glucanase SdGluc5_26A from Saccharophagus degradans [Saccharophagus degradans 2-40] |
| 4XZW_A | 4.23e-16 | 35 | 333 | 12 | 302 | Endo-glucanasechimera C10 [uncultured bacterium] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P19570 | 2.63e-14 | 60 | 327 | 94 | 398 | Endoglucanase C OS=Evansella cellulosilytica (strain ATCC 21833 / DSM 2522 / FERM P-1141 / JCM 9156 / N-4) OX=649639 GN=celC PE=3 SV=1 |
| P22541 | 2.35e-13 | 35 | 304 | 116 | 369 | Endoglucanase A OS=Butyrivibrio fibrisolvens OX=831 GN=celA PE=1 SV=1 |
| P15704 | 2.68e-11 | 35 | 333 | 47 | 335 | Endoglucanase OS=Clostridium saccharobutylicum OX=169679 GN=eglA PE=3 SV=1 |
| P19424 | 3.97e-10 | 60 | 304 | 256 | 519 | Endoglucanase OS=Bacillus sp. (strain KSM-635) OX=1415 PE=1 SV=1 |
| A0A0U4EBH5 | 4.19e-10 | 54 | 304 | 72 | 328 | Cellulase CelDZ1 OS=Thermoanaerobacterium sp. OX=40549 GN=celDZ1a PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
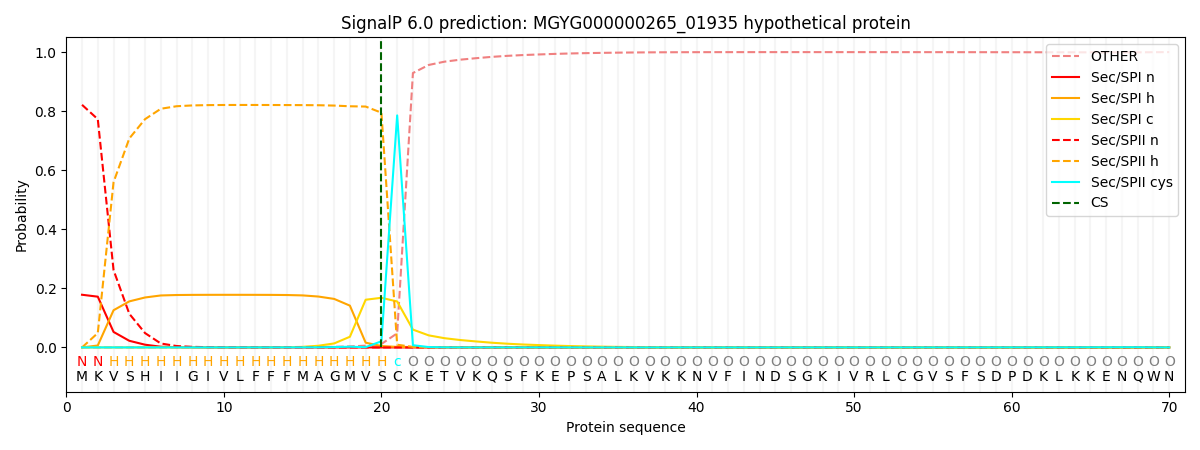
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000487 | 0.174583 | 0.824679 | 0.000094 | 0.000077 | 0.000067 |
