You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000272_02659
You are here: Home > Sequence: MGYG000000272_02659
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
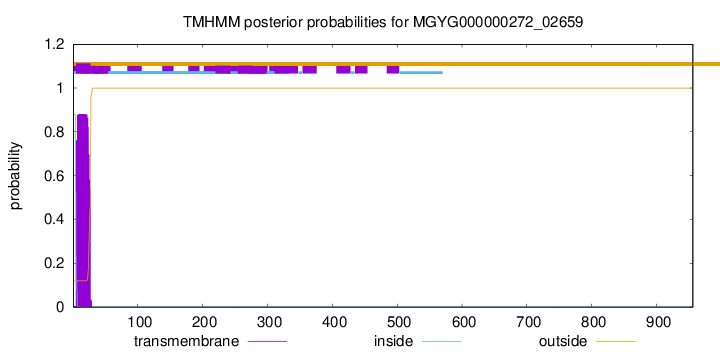
TMHMM annotations
Basic Information help
| Species | Prevotella hominis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Prevotella; Prevotella hominis | |||||||||||
| CAZyme ID | MGYG000000272_02659 | |||||||||||
| CAZy Family | PL1 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 3204; End: 6074 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| PL1 | 463 | 636 | 4.7e-41 | 0.8465346534653465 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3866 | PelB | 1.18e-47 | 331 | 749 | 1 | 345 | Pectate lyase [Carbohydrate transport and metabolism]. |
| pfam18283 | CBM77 | 4.99e-30 | 839 | 949 | 1 | 108 | Carbohydrate binding module 77. This domain is the non-catalytic carbohydrate binding module 77 (CBM77) present in Ruminococcus flavefaciens. CBMs fulfil a critical targeting function in plant cell wall depolymerisation. In CBM77, a cluster of conserved basic residues (Lys1092, Lys1107 and Lys1162) confer calcium-independent recognition of homogalacturonan. |
| smart00656 | Amb_all | 4.18e-29 | 471 | 637 | 17 | 190 | Amb_all domain. |
| pfam00544 | Pec_lyase_C | 4.47e-16 | 435 | 633 | 1 | 211 | Pectate lyase. This enzyme forms a right handed beta helix structure. Pectate lyase is an enzyme involved in the maceration and soft rotting of plant tissue. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QNT67442.1 | 0.0 | 189 | 956 | 17 | 778 |
| QUT73893.1 | 4.35e-227 | 277 | 953 | 20 | 683 |
| QCD40816.1 | 1.12e-210 | 218 | 789 | 7 | 588 |
| QCP73706.1 | 1.12e-210 | 218 | 789 | 7 | 588 |
| QOR20273.1 | 4.52e-194 | 280 | 782 | 50 | 552 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3VMV_A | 2.33e-28 | 462 | 706 | 72 | 317 | Crystalstructure of pectate lyase Bsp165PelA from Bacillus sp. N165 [Bacillus sp. N16-5],3VMW_A Crystal structure of pectate lyase Bsp165PelA from Bacillus sp. N165 in complex with trigalacturonate [Bacillus sp. N16-5] |
| 1VBL_A | 1.21e-18 | 471 | 633 | 133 | 330 | Structureof the thermostable pectate lyase PL 47 [Bacillus sp. TS-47] |
| 1AIR_A | 7.14e-15 | 419 | 747 | 48 | 342 | ChainA, PECTATE LYASE C [Dickeya chrysanthemi],1O88_A Chain A, Pectate Lyase C [Dickeya chrysanthemi],1O8D_A Chain A, PECTATE LYASE C [Dickeya chrysanthemi],1O8E_A Chain A, PECTATE LYASE C [Dickeya chrysanthemi],1O8F_A Chain A, PECTATE LYASE C [Dickeya chrysanthemi],1O8G_A Chain A, PECTATE LYASE C [Dickeya chrysanthemi],1O8H_A Chain A, PECTATE LYASE C [Dickeya chrysanthemi],1O8I_A Chain A, PECTATE LYASE C [Dickeya chrysanthemi],1O8J_A Chain A, PECTATE LYASE C [Dickeya chrysanthemi],1O8K_A Chain A, PECTATE LYASE C [Dickeya chrysanthemi],1O8L_A Chain A, PECTATE LYASE C [Dickeya chrysanthemi],1O8M_A Chain A, PECTATE LYASE C [Dickeya chrysanthemi],1PLU_A Chain A, Protein (pectate Lyase C) [Dickeya chrysanthemi],2PEC_A Chain A, PECTATE LYASE C [Dickeya chrysanthemi] |
| 3ZSC_A | 8.36e-15 | 479 | 610 | 76 | 213 | Catalyticfunction and substrate recognition of the pectate lyase from Thermotoga maritima [Thermotoga maritima] |
| 2EWE_A | 1.71e-14 | 419 | 747 | 48 | 342 | ChainA, Pectate lyase C [Dickeya chrysanthemi] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| B1B6T1 | 7.41e-21 | 412 | 642 | 60 | 281 | Pectate trisaccharide-lyase OS=Bacillus sp. OX=1409 GN=pel PE=1 SV=1 |
| Q65DC2 | 7.41e-21 | 412 | 642 | 60 | 281 | Pectate trisaccharide-lyase OS=Bacillus licheniformis (strain ATCC 14580 / DSM 13 / JCM 2505 / CCUG 7422 / NBRC 12200 / NCIMB 9375 / NCTC 10341 / NRRL NRS-1264 / Gibson 46) OX=279010 GN=BLi04129 PE=3 SV=1 |
| Q8GCB2 | 7.41e-21 | 412 | 642 | 60 | 281 | Pectate trisaccharide-lyase OS=Bacillus licheniformis OX=1402 GN=pelA PE=1 SV=1 |
| Q5AVN4 | 1.55e-15 | 471 | 646 | 99 | 277 | Pectate lyase A OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=plyA PE=1 SV=1 |
| P11073 | 4.80e-14 | 419 | 747 | 70 | 364 | Pectate lyase C OS=Dickeya chrysanthemi OX=556 GN=pelC PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
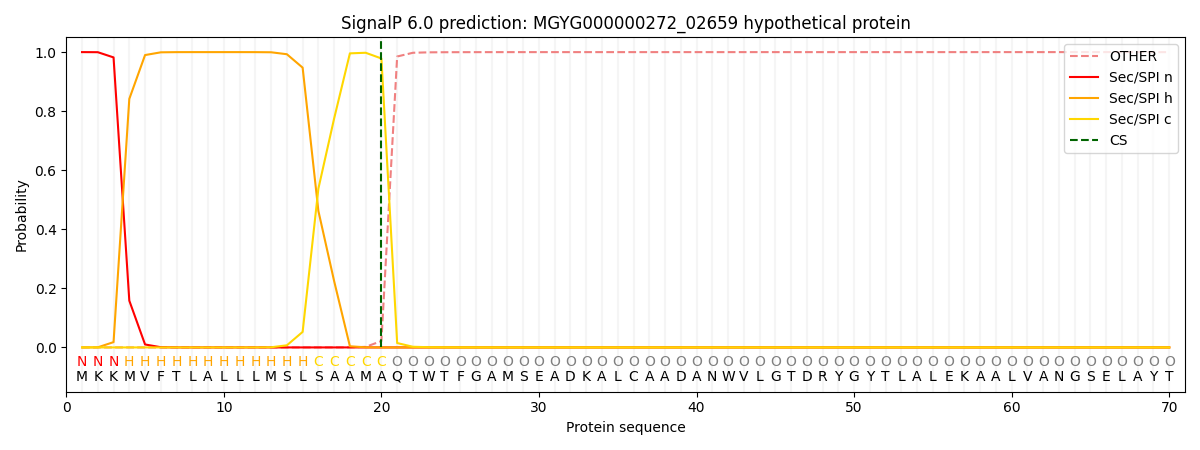
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000255 | 0.999060 | 0.000183 | 0.000178 | 0.000158 | 0.000146 |