You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000273_02005
You are here: Home > Sequence: MGYG000000273_02005
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
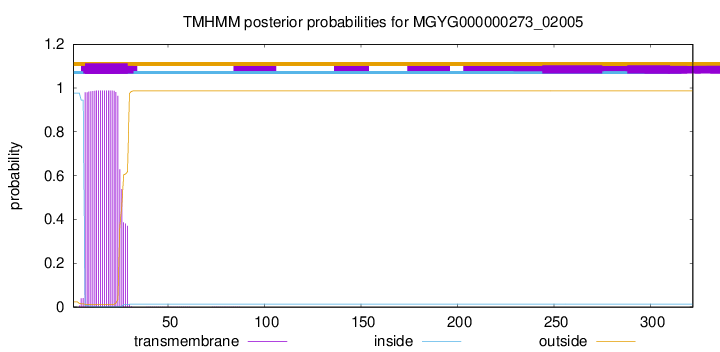
TMHMM annotations
Basic Information help
| Species | Phocaeicola coprophilus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Phocaeicola; Phocaeicola coprophilus | |||||||||||
| CAZyme ID | MGYG000000273_02005 | |||||||||||
| CAZy Family | GH23 | |||||||||||
| CAZyme Description | Membrane-bound lytic murein transglycosylase D | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 56827; End: 57795 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH23 | 93 | 229 | 2.9e-27 | 0.9111111111111111 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd16894 | MltD-like | 3.59e-65 | 101 | 226 | 1 | 129 | Membrane-bound lytic murein transglycosylase D and similar proteins. Lytic transglycosylases (LT) catalyze the cleavage of the beta-1,4-glycosidic bond between N-acetylmuramic acid (MurNAc) and N-acetyl-D-glucosamine (GlcNAc). Membrane-bound lytic murein transglycosylase D protein (MltD) family members may have one or more small LysM domains, which may contribute to peptidoglycan binding. Unlike the similar "goose-type" lysozymes, LTs also make a new glycosidic bond with the C6 hydroxyl group of the same muramic acid residue. Proteins similar to this family include the soluble and insoluble membrane-bound LTs in bacteria, the LTs in bacteriophage lambda, as well as the eukaryotic "goose-type" lysozymes (goose egg-white lysozyme; GEWL). |
| PRK10783 | mltD | 2.72e-28 | 66 | 242 | 81 | 258 | membrane-bound lytic murein transglycosylase D; Provisional |
| pfam01464 | SLT | 9.11e-26 | 96 | 202 | 1 | 108 | Transglycosylase SLT domain. This family is distantly related to pfam00062. Members are found in phages, type II, type III and type IV secretion systems. |
| cd00254 | LT-like | 3.73e-24 | 113 | 224 | 7 | 109 | lytic transglycosylase(LT)-like domain. Members include the soluble and insoluble membrane-bound LTs in bacteria and LTs in bacteriophage lambda. LTs catalyze the cleavage of the beta-1,4-glycosidic bond between N-acetylmuramic acid (MurNAc) and N-acetyl-D-glucosamine (GlcNAc), as do "goose-type" lysozymes. However, in addition to this, they also make a new glycosidic bond with the C6 hydroxyl group of the same muramic acid residue. |
| cd13401 | Slt70-like | 1.50e-18 | 93 | 223 | 6 | 144 | 70kDa soluble lytic transglycosylase (Slt70) and similar proteins. Catalytic domain of the 70kda soluble lytic transglycosylase (LT)-like proteins, which also have an N-terminal U-shaped U-domain and a linker L-domain. LTs catalyze the cleavage of the beta-1,4-glycosidic bond between N-acetylmuramic acid (MurNAc) and N-acetyl-D-glucosamine (GlcNAc), as do "goose-type" lysozymes. However, in addition to this, they also make a new glycosidic bond with the C6 hydroxyl group of the same muramic acid residue. Proteins similar to this family include the soluble and insoluble membrane-bound LTs in bacteria and the LTs in bacteriophage lambda. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QRO24006.1 | 1.59e-242 | 1 | 322 | 1 | 322 |
| ADY36030.1 | 9.31e-155 | 12 | 320 | 15 | 322 |
| QUT56374.1 | 3.30e-145 | 16 | 322 | 16 | 332 |
| QQY40386.1 | 1.34e-144 | 16 | 322 | 16 | 332 |
| ABR38313.1 | 1.71e-144 | 16 | 322 | 23 | 339 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5OHU_A | 9.49e-07 | 117 | 261 | 503 | 642 | TheX-ray Structure of Lytic Transglycosylase Slt from Pseudomonas aeruginosa [Pseudomonas aeruginosa] |
| 6FBT_A | 1.24e-06 | 117 | 223 | 474 | 587 | ChainA, Lytic murein transglycosylase [Pseudomonas aeruginosa] |
| 6FC4_A | 2.91e-06 | 117 | 223 | 475 | 588 | ChainA, Soluble lytic murein transglycosylase [Pseudomonas aeruginosa] |
| 6FCQ_A | 2.91e-06 | 117 | 223 | 474 | 587 | ChainA, Soluble lytic murein transglycosylase [Pseudomonas aeruginosa],6FCR_A Chain A, Soluble lytic murein transglycosylase [Pseudomonas aeruginosa],6FCS_A Chain A, Soluble lytic murein transglycosylase [Pseudomonas aeruginosa],6FCU_A Chain A, Soluble lytic murein transglycosylase [Pseudomonas aeruginosa] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P0AEZ7 | 1.51e-26 | 43 | 302 | 52 | 310 | Membrane-bound lytic murein transglycosylase D OS=Escherichia coli (strain K12) OX=83333 GN=mltD PE=1 SV=1 |
| P0AEZ8 | 1.51e-26 | 43 | 302 | 52 | 310 | Membrane-bound lytic murein transglycosylase D OS=Escherichia coli O6:H1 (strain CFT073 / ATCC 700928 / UPEC) OX=199310 GN=mltD PE=3 SV=1 |
| P32820 | 6.70e-14 | 89 | 199 | 21 | 132 | Putative tributyltin chloride resistance protein OS=Alteromonas sp. (strain M-1) OX=29457 GN=tbtA PE=3 SV=1 |
| O31976 | 6.93e-08 | 117 | 223 | 1447 | 1539 | SPbeta prophage-derived uncharacterized transglycosylase YomI OS=Bacillus subtilis (strain 168) OX=224308 GN=yomI PE=3 SV=2 |
| O64046 | 6.93e-08 | 117 | 223 | 1447 | 1539 | Probable tape measure protein OS=Bacillus phage SPbeta OX=66797 GN=yomI PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
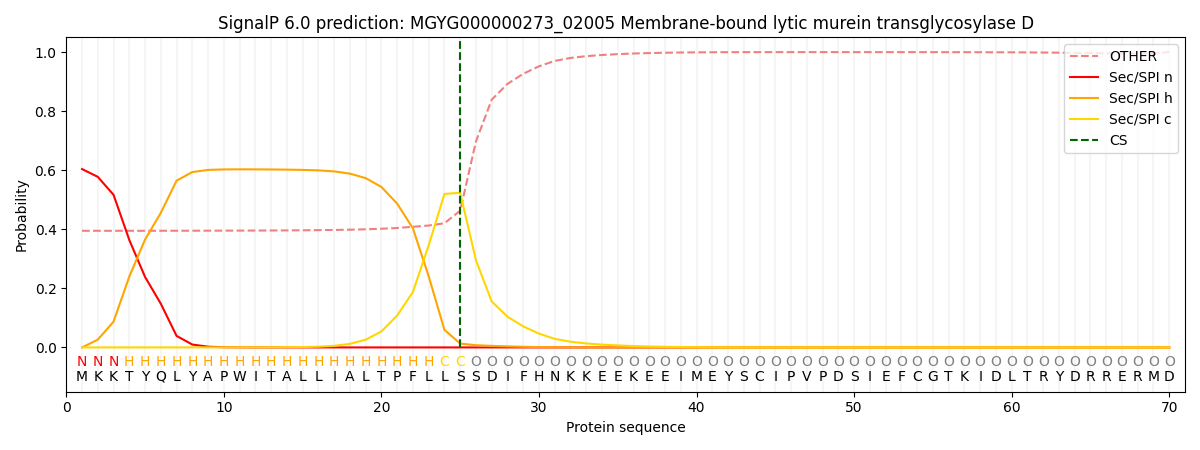
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.421378 | 0.573690 | 0.002531 | 0.000913 | 0.000550 | 0.000920 |