You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000273_02446
You are here: Home > Sequence: MGYG000000273_02446
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Phocaeicola coprophilus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Phocaeicola; Phocaeicola coprophilus | |||||||||||
| CAZyme ID | MGYG000000273_02446 | |||||||||||
| CAZy Family | GH20 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 5155; End: 6975 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH20 | 147 | 468 | 4.4e-44 | 0.9584569732937686 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd06565 | GH20_GcnA-like | 1.53e-81 | 149 | 467 | 1 | 301 | Glycosyl hydrolase family 20 (GH20) catalytic domain of N-acetyl-beta-D-glucosaminidase (GcnA, also known as BhsA) and related proteins. GcnA is an exoglucosidase which cleaves N-acetyl-beta-D-galactosamine (NAG) and N-acetyl-beta-D-galactosamine residues from 4-methylumbelliferylated (4MU) substrates, as well as cleaving NAG from chito-oligosaccharides (i.e. NAG polymers). In contrast, sulfated forms of the substrate are unable to be cleaved and act instead as mild competitive inhibitors. Additionally, the enzyme is known to be poisoned by several first-row transition metals as well as by mercury. GcnA forms a homodimer with subunits comprised of three domains, an N-terminal zincin-like domain, this central catalytic GH20 domain, and a C-terminal alpha helical domain. The GH20 hexosaminidases are thought to act via a catalytic mechanism in which the catalytic nucleophile is not provided by solvent or the enzyme, but by the substrate itself. |
| COG3525 | Chb | 4.19e-23 | 84 | 330 | 196 | 488 | N-acetyl-beta-hexosaminidase [Carbohydrate transport and metabolism]. |
| pfam00728 | Glyco_hydro_20 | 3.04e-22 | 148 | 366 | 2 | 250 | Glycosyl hydrolase family 20, catalytic domain. This domain has a TIM barrel fold. |
| cd06563 | GH20_chitobiase-like | 5.20e-21 | 148 | 330 | 2 | 226 | The chitobiase of Serratia marcescens is a beta-N-1,4-acetylhexosaminidase with a glycosyl hydrolase family 20 (GH20) domain that hydrolyzes the beta-1,4-glycosidic linkages in oligomers derived from chitin. Chitin is degraded by a two step process: i) a chitinase hydrolyzes the chitin to oligosaccharides and disaccharides such as di-N-acetyl-D-glucosamine and chitobiose, ii) chitobiase then further degrades these oligomers into monomers. This GH20 domain family includes an N-acetylglucosamidase (GlcNAcase A) from Pseudoalteromonas piscicida and an N-acetylhexosaminidase (SpHex) from Streptomyces plicatus. SpHex lacks the C-terminal PKD (polycystic kidney disease I)-like domain found in the chitobiases. The GH20 hexosaminidases are thought to act via a catalytic mechanism in which the catalytic nucleophile is not provided by solvent or the enzyme, but by the substrate itself. |
| cd02742 | GH20_hexosaminidase | 6.03e-21 | 149 | 331 | 1 | 193 | Beta-N-acetylhexosaminidases of glycosyl hydrolase family 20 (GH20) catalyze the removal of beta-1,4-linked N-acetyl-D-hexosamine residues from the non-reducing ends of N-acetyl-beta-D-hexosaminides including N-acetylglucosides and N-acetylgalactosides. These enzymes are broadly distributed in microorganisms, plants and animals, and play roles in various key physiological and pathological processes. These processes include cell structural integrity, energy storage, cellular signaling, fertilization, pathogen defense, viral penetration, the development of carcinomas, inflammatory events and lysosomal storage disorders. The GH20 enzymes include the eukaryotic beta-N-acetylhexosaminidases A and B, the bacterial chitobiases, dispersin B, and lacto-N-biosidase. The GH20 hexosaminidases are thought to act via a catalytic mechanism in which the catalytic nucleophile is not provided by the solvent or the enzyme, but by the substrate itself. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QRO25975.1 | 0.0 | 1 | 579 | 1 | 579 |
| QUT49641.1 | 3.51e-297 | 3 | 579 | 2 | 578 |
| QQY42843.1 | 6.69e-283 | 27 | 579 | 26 | 578 |
| QQY39583.1 | 6.69e-283 | 27 | 579 | 26 | 578 |
| ABR38110.1 | 6.69e-283 | 27 | 579 | 26 | 578 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6YHH_A | 6.27e-23 | 65 | 390 | 66 | 408 | X-rayStructure of Flavobacterium johnsoniae chitobiase (FjGH20) [Flavobacterium johnsoniae UW101],6YHH_B X-ray Structure of Flavobacterium johnsoniae chitobiase (FjGH20) [Flavobacterium johnsoniae UW101] |
| 6Q63_A | 7.49e-23 | 77 | 381 | 90 | 436 | BT0459[Bacteroides thetaiotaomicron],6Q63_B BT0459 [Bacteroides thetaiotaomicron],6Q63_C BT0459 [Bacteroides thetaiotaomicron] |
| 4PYS_A | 1.03e-20 | 86 | 334 | 70 | 347 | Thecrystal structure of beta-N-acetylhexosaminidase from Bacteroides fragilis NCTC 9343 [Bacteroides fragilis NCTC 9343],4PYS_B The crystal structure of beta-N-acetylhexosaminidase from Bacteroides fragilis NCTC 9343 [Bacteroides fragilis NCTC 9343] |
| 1M04_A | 1.42e-20 | 28 | 468 | 26 | 476 | ChainA, Beta-N-acetylhexosaminidase [Streptomyces plicatus] |
| 1M03_A | 1.42e-20 | 28 | 468 | 26 | 476 | ChainA, Beta-N-acetylhexosaminidase [Streptomyces plicatus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q7WUL4 | 4.79e-15 | 24 | 303 | 1 | 315 | Beta-N-acetylhexosaminidase OS=Cellulomonas fimi OX=1708 GN=hex20 PE=1 SV=1 |
| A6QNR0 | 3.37e-14 | 150 | 444 | 2 | 290 | Hexosaminidase D OS=Bos taurus OX=9913 GN=HEXD PE=2 SV=2 |
| Q3U4H6 | 7.10e-12 | 150 | 444 | 10 | 298 | Hexosaminidase D OS=Mus musculus OX=10090 GN=Hexd PE=1 SV=1 |
| Q8WVB3 | 2.18e-11 | 150 | 444 | 10 | 298 | Hexosaminidase D OS=Homo sapiens OX=9606 GN=HEXD PE=1 SV=3 |
| Q04786 | 9.31e-10 | 77 | 237 | 247 | 438 | Beta-hexosaminidase OS=Vibrio vulnificus OX=672 GN=hex PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
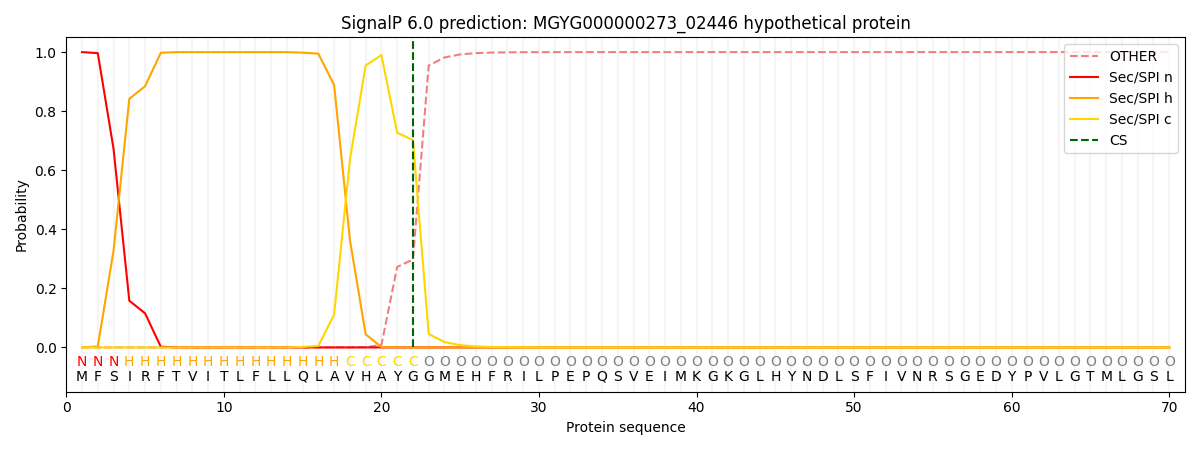
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000225 | 0.999215 | 0.000150 | 0.000141 | 0.000132 | 0.000127 |
